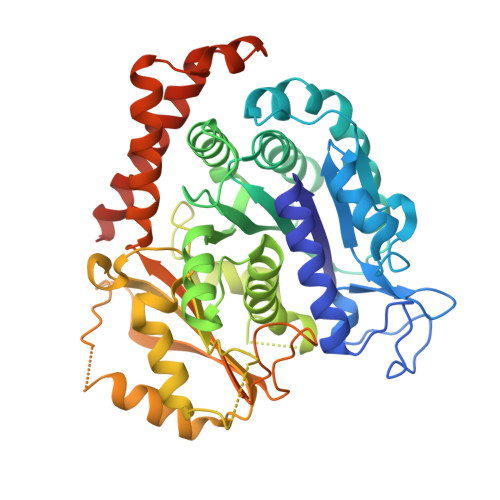
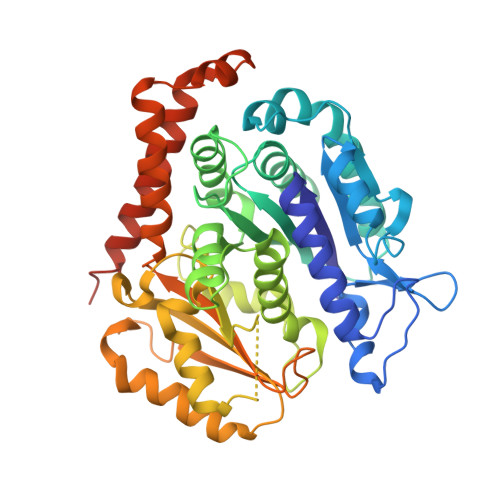
A novel orally active microtubule destabilizing agent S-40 targets the colchicine-binding site and shows potent antitumor activity.
Du, T., Lin, S., Ji, M., Xue, N., Liu, Y., Zhang, Z., Zhang, K., Zhang, J., Zhang, Y., Wang, Q., Sheng, L., Li, Y., Lu, D., Chen, X., Xu, H.(2020) Cancer Lett 495: 22-32
- PubMed: 32931884
- DOI: https://doi.org/10.1016/j.canlet.2020.08.040
- Primary Citation of Related Structures:
6LS4 - PubMed Abstract:
The tubulin colchicine binding site has been recognized as an attractive drug target to combat cancer, but none of the candidate drugs have been approved for medical treatment. We recently identified a structurally distinct small molecule S-40 as an oral potent tubulin destabilizing agent. Crystal structure analysis of S-40 in a complex with tubulin at a resolution of 2.4 Å indicated that S-40 occupies all 3 zones in the colchicine pocket with interactions different from known microtubule inhibitors, presenting unique effects on assembly and curvature of tubulin dimers. S-40 overcomes paclitaxel resistance and lacks neurotoxicity, which are the main obstacles limiting clinical applications of paclitaxel. Moreover, S-40 harbors the ability to inhibit growth of cancer cell lines as well as patient-derived organoids, induce mitotic arrest and cell apoptosis. Xenograft mouse models of human prostate cancer DU145, non-small cell lung cancer NCI-H1299 and paclitaxel-resistant A549 were strongly restrained without apparent side effects by S-40 oral administration once daily. These findings provide evidence for the development of S-40 as the next generation of orally effective microtubule inhibitors for cancer therapy.
Organizational Affiliation:
State Key Laboratory of Bioactive Substance and Function of Natural Medicines, Institute of Materia Medica, Chinese Academy of Medical Sciences and Peking Union Medical College, Beijing, 100050, PR China.