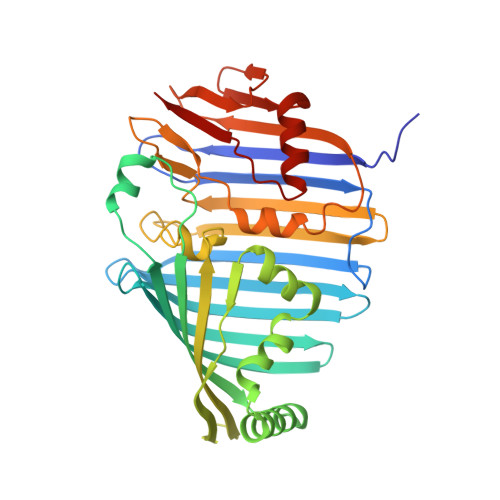
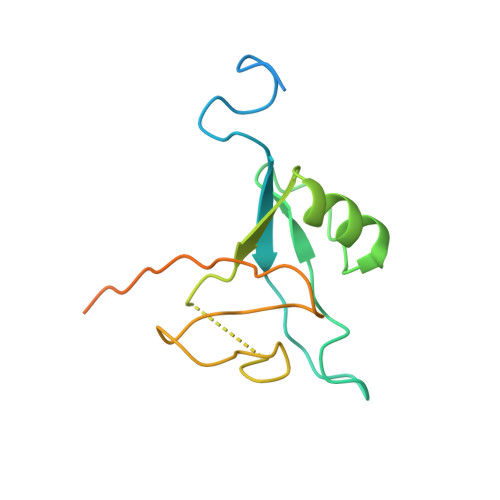
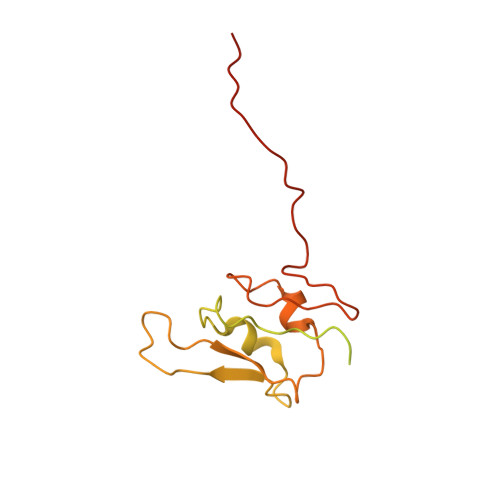
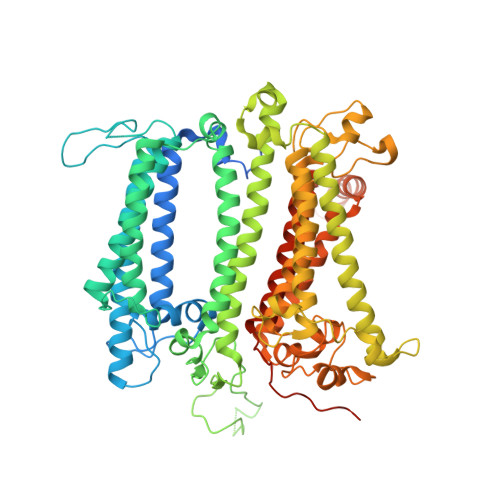
Architecture of the photosynthetic complex from a green sulfur bacterium.
Chen, J.H., Wu, H., Xu, C., Liu, X.C., Huang, Z., Chang, S., Wang, W., Han, G., Kuang, T., Shen, J.R., Zhang, X.(2020) Science 370
- PubMed: 33214250
- DOI: https://doi.org/10.1126/science.abb6350
- Primary Citation of Related Structures:
6M32 - PubMed Abstract:
The photosynthetic apparatus of green sulfur bacteria (GSB) contains a peripheral antenna chlorosome, light-harvesting Fenna-Matthews-Olson proteins (FMO), and a reaction center (GsbRC). We used cryo-electron microscopy to determine a 2.7-angstrom structure of the FMO-GsbRC supercomplex from Chlorobaculum tepidum The GsbRC binds considerably fewer (bacterio)chlorophylls [(B)Chls] than other known type I RCs do, and the organization of (B)Chls is similar to that in photosystem II. Two BChl layers in GsbRC are not connected by Chls, as seen in other RCs, but associate with two carotenoid derivatives. Relatively long distances of 22 to 33 angstroms were observed between BChls of FMO and GsbRC, consistent with the inefficient energy transfer between these entities. The structure contains common features of both type I and type II RCs and provides insight into the evolution of photosynthetic RCs.
Organizational Affiliation:
Department of Pathology of Sir Run Run Shaw Hospital and Department of Biophysics, Zhejiang University School of Medicine, Hangzhou, 310058 Zhejiang, China.