An orally available M pro inhibitor is effective against wild-type SARS-CoV-2 and variants including Omicron.
Quan, B.X., Shuai, H., Xia, A.J., Hou, Y., Zeng, R., Liu, X.L., Lin, G.F., Qiao, J.X., Li, W.P., Wang, F.L., Wang, K., Zhou, R.J., Yuen, T.T., Chen, M.X., Yoon, C., Wu, M., Zhang, S.Y., Huang, C., Wang, Y.F., Yang, W., Tian, C., Li, W.M., Wei, Y.Q., Yuen, K.Y., Chan, J.F., Lei, J., Chu, H., Yang, S.(2022) Nat Microbiol 7: 716-725
- PubMed: 35477751
- DOI: https://doi.org/10.1038/s41564-022-01119-7
- Primary Citation of Related Structures:
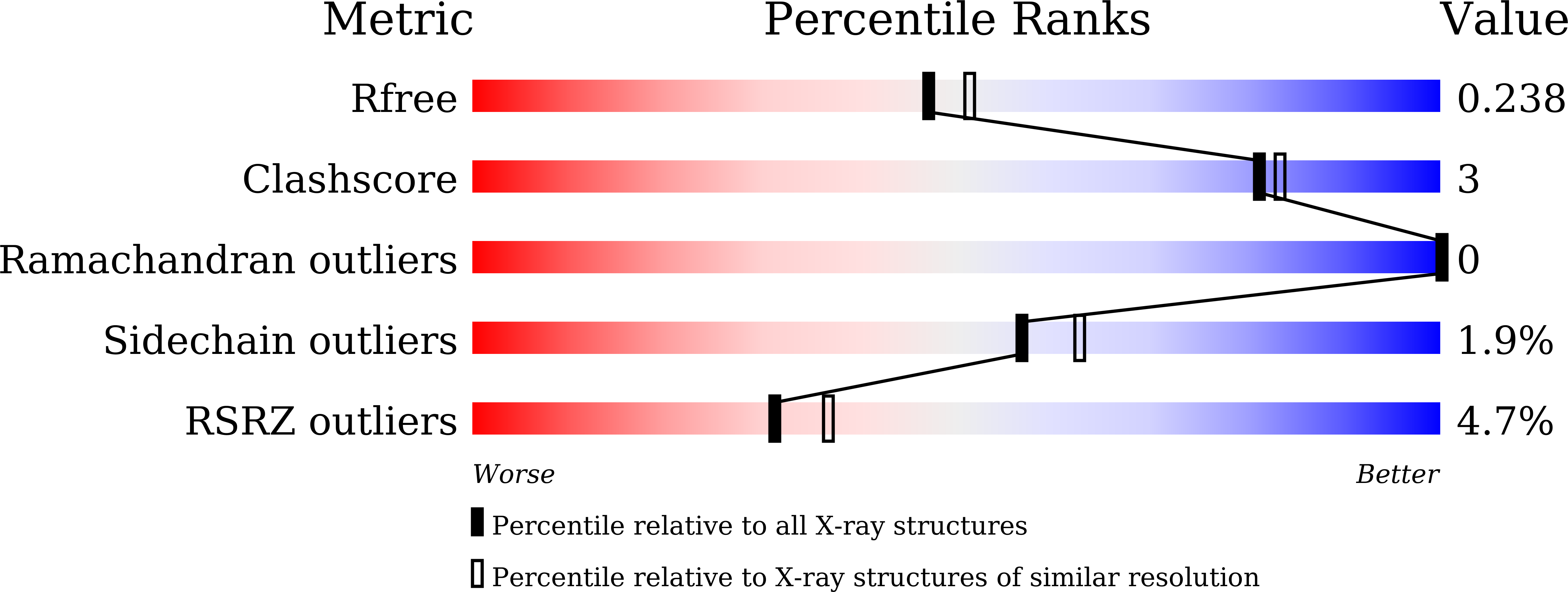
7FAY, 7FAZ - PubMed Abstract:
Emerging SARS-CoV-2 variants continue to cause waves of new infections globally. Developing effective antivirals against SARS-CoV-2 and its variants is an urgent task. The main protease (M pro ) of SARS-CoV-2 is an attractive drug target because of its central role in viral replication and its conservation among variants. We herein report a series of potent α-ketoamide-containing M pro inhibitors obtained using the Ugi four-component reaction. The prioritized compound, Y180, showed an IC 50 of 8.1 nM against SARS-CoV-2 M pro and had oral bioavailability of 92.9%, 31.9% and 85.7% in mice, rats and dogs, respectively. Y180 protected against wild-type SARS-CoV-2, B.1.1.7 (Alpha), B.1.617.1 (Kappa) and P.3 (Theta), with EC 50 of 11.4, 20.3, 34.4 and 23.7 nM, respectively. Oral treatment with Y180 displayed a remarkable antiviral potency and substantially ameliorated the virus-induced tissue damage in both nasal turbinate and lung of B.1.1.7-infected K18-human ACE2 (K18-hACE2) transgenic mice. Therapeutic treatment with Y180 improved the survival of mice from 0 to 44.4% (P = 0.0086) upon B.1.617.1 infection in the lethal infection model. Importantly, Y180 was also highly effective against the B.1.1.529 (Omicron) variant both in vitro and in vivo. Overall, our study provides a promising lead compound for oral drug development against SARS-CoV-2.
Organizational Affiliation:
State Key Laboratory of Biotherapy and Cancer Center, West China Hospital, Sichuan University, Chengdu, Sichuan, China.