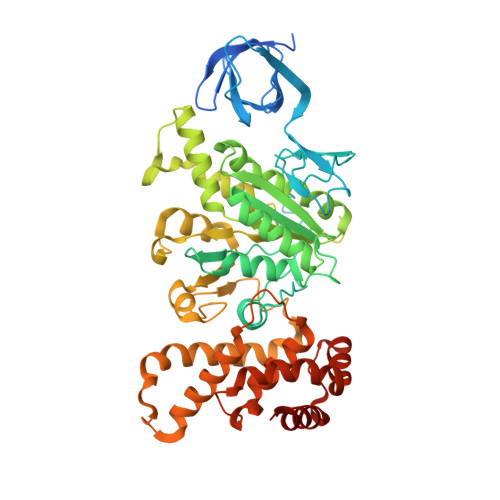
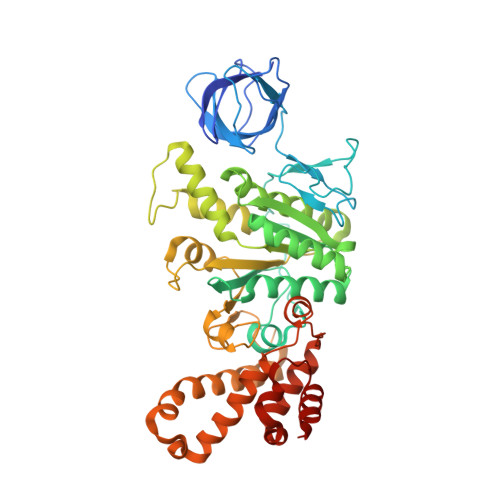
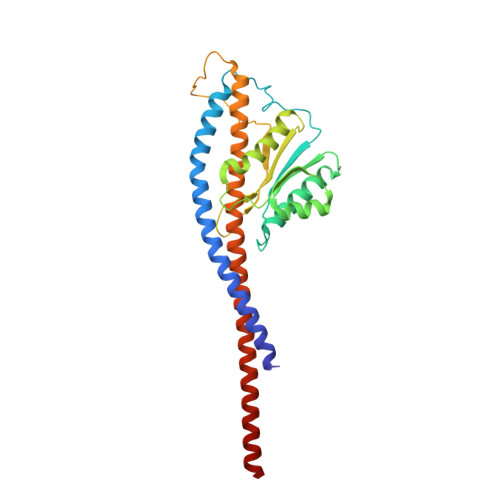
The six steps of the complete F 1 -ATPase rotary catalytic cycle.
Sobti, M., Ueno, H., Noji, H., Stewart, A.G.(2021) Nat Commun 12: 4690-4690
- PubMed: 34344897
- DOI: https://doi.org/10.1038/s41467-021-25029-0
- Primary Citation of Related Structures:
7L1Q, 7L1R, 7L1S - PubMed Abstract:
F 1 F o ATP synthase interchanges phosphate transfer energy and proton motive force via a rotary catalysis mechanism. Isolated F 1 -ATPase catalytic cores can hydrolyze ATP, passing through six intermediate conformational states to generate rotation of their central γ-subunit. Although previous structural studies have contributed greatly to understanding rotary catalysis in the F 1 -ATPase, the structure of an important conformational state (the binding-dwell) has remained elusive. Here, we exploit temperature and time-resolved cryo-electron microscopy to determine the structure of the binding- and catalytic-dwell states of Bacillus PS3 F 1 -ATPase. Each state shows three catalytic β-subunits in different conformations, establishing the complete set of six states taken up during the catalytic cycle and providing molecular details for both the ATP binding and hydrolysis strokes. We also identify a potential phosphate-release tunnel that indicates how ADP and phosphate binding are coordinated during synthesis. Overall these findings provide a structural basis for the entire F 1 -ATPase catalytic cycle.
Organizational Affiliation:
Molecular, Structural and Computational Biology Division, The Victor Chang Cardiac Research Institute, Darlinghurst, NSW, Australia.