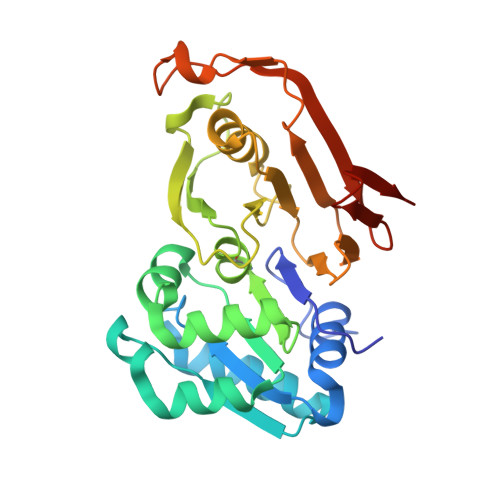
The structure of cyanophycinase in complex with a cyanophycin degradation intermediate.
Sharon, I., Grogg, M., Hilvert, D., Schmeing, T.M.(2022) Biochim Biophys Acta Gen Subj 1866: 130217-130217
- PubMed: 35905922
- DOI: https://doi.org/10.1016/j.bbagen.2022.130217
- Primary Citation of Related Structures:
7UQV, 7UQW - PubMed Abstract:
Cyanophycinases are serine protease family enzymes which are required for the metabolism of cyanophycin, the natural polymer multi-L-arginyl-poly(L-aspartic acid). Cyanophycinases degrade cyanophycin to β-Asp-Arg dipeptides, which enables use of this important store of fixed nitrogen.
Organizational Affiliation:
Department of Biochemistry and Centre de recherche en biologie structurale, McGill University, Montréal, QC H3G 0B1, Canada.