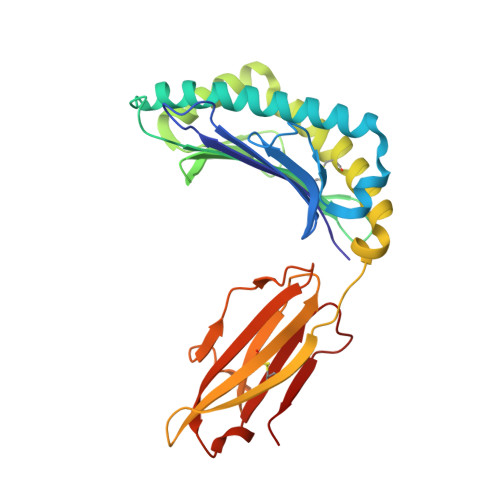
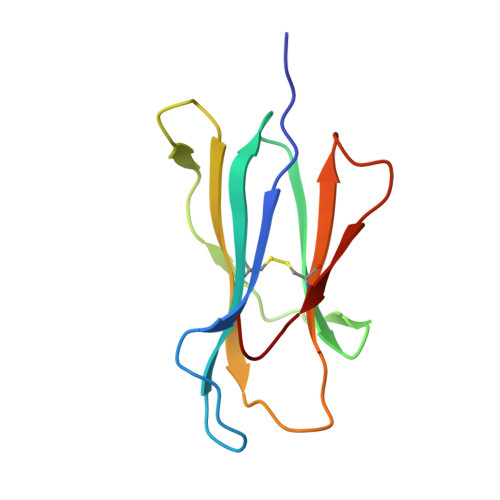
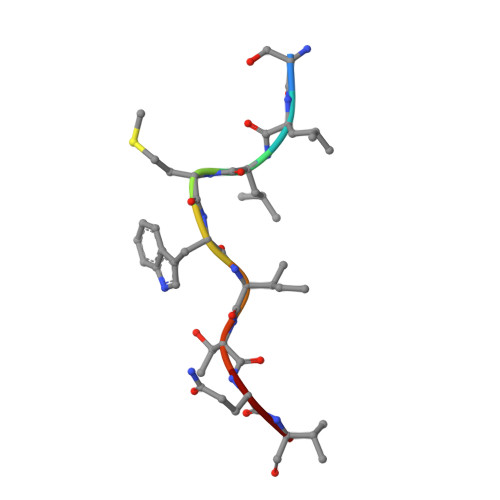
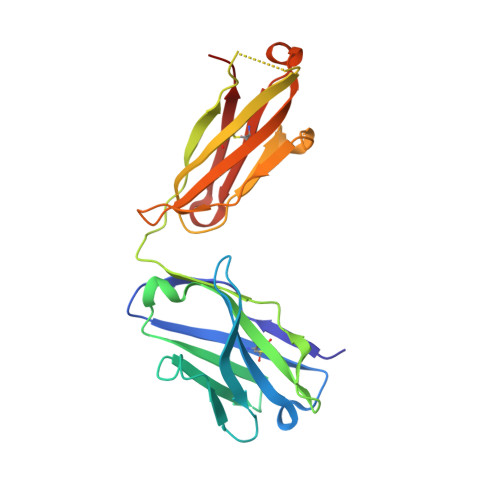
HLA-A∗02-gated safety switch for cancer therapy has exquisite specificity for its allelic target antigen.
Mock, J.Y., Winters, A., Riley, T.P., Bruno, R., Naradikian, M.S., Sharma, S., Jette, C.A., Elshimali, R., Gahrs, C., Toledo-Warshaviak, D., West Jr., A.P., Kamb, A., Hamburger, A.E.(2022) Mol Ther Oncolytics 27: 157-166
- PubMed: 36381658
- DOI: https://doi.org/10.1016/j.omto.2022.09.010
- Primary Citation of Related Structures:
8EB2 - PubMed Abstract:
Innovative cell-based therapies are important new weapons in the fight against difficult-to-treat cancers. One promising strategy involves cell therapies equipped with multiple receptors to integrate signals from more than one antigen. We developed a specific embodiment of this approach called Tmod, a two-receptor system that combines activating and inhibitory inputs to distinguish between tumor and normal cells. The selectivity of Tmod is enforced by the inhibitory receptor (blocker) that recognizes an antigen, such as an HLA allele, whose expression is absent from tumors because of loss of heterozygosity. Although unwanted cross-reactivity of the blocker likely reduces efficacy rather than safety, it is important to verify the blocker's specificity. We have tested an A∗02-directed blocker derived from the PA2.1 mouse antibody as a safety mechanism paired with a mesothelin-specific activating CAR in our Tmod construct. We solved the crystal structure of humanized PA2.1 Fab in complex with HLA-A∗02 to determine its binding epitope, which was used to bioinformatically select specific class I HLA alleles to test the blocker's functional specificity in vitro . We found that this A∗02-directed blocker is highly specific for its cognate antigen, with only one cross-reactive allele (A∗69) capable of triggering comparable function.
Organizational Affiliation:
A2 Biotherapeutics, 30301 Agoura Road, Agoura Hills, CA 91301, USA.