Full Text |
QUERY: Chemical ID(s) = KGM | MyPDB Login | Search API |
| Search Summary | This query matches 10 Structures. |
Structure Determination MethodologyScientific Name of Source OrganismTaxonomyExperimental MethodPolymer Entity TypeRefinement Resolution (Å)Release DateEnzyme Classification NameMembrane Protein AnnotationSymmetry TypeSCOP Classification | 1 to 10 of 10 Structures Page 1 of 1 Sort by
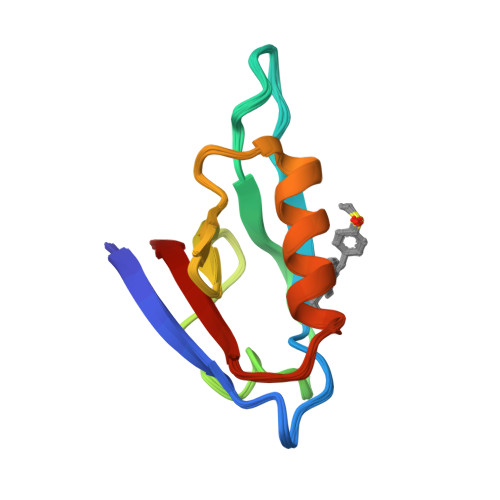
NMR structure of the mDvl1 PDZ domain in complex with its inhibitorLee, H.J., Shao, Y., Wang, N.X., Shi, D.L., Zheng, J.J. (2009) Angew Chem Int Ed Engl 48: 6448-6452
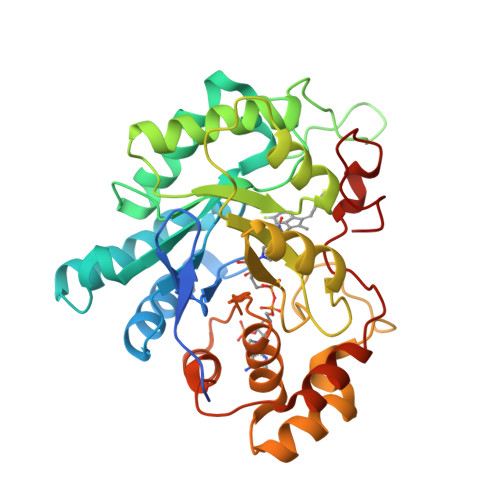
AKR1C3 complex with sulindacYosaatmadja, Y., Teague, R.M., Flanagan, J.U., Squire, C.J. (2012) PLoS One 7: e43965-e43965
Crystal Structure of Human Aldose Reductase Complexed with SulindacZheng, X., Chen, J., Luo, H., Hu, X. (2012) FEBS Lett 586: 55-59
Aldose reductase in complex with NSAID-type inhibitor at 1.0 A resolution(2011) ChemMedChem 6: 2155-2157
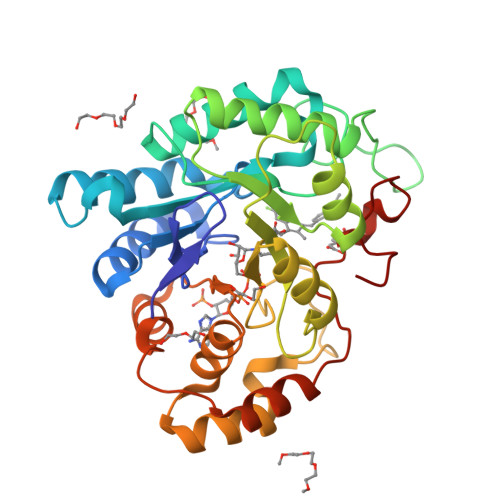
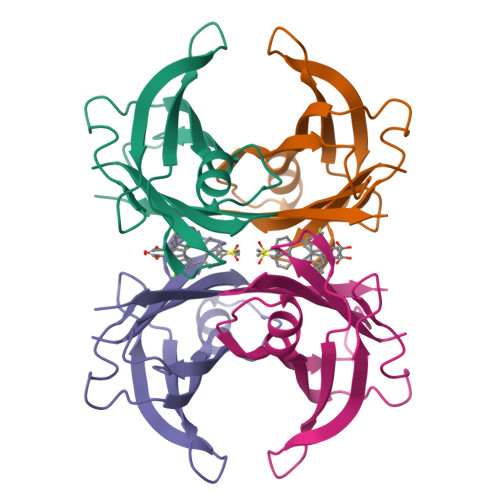
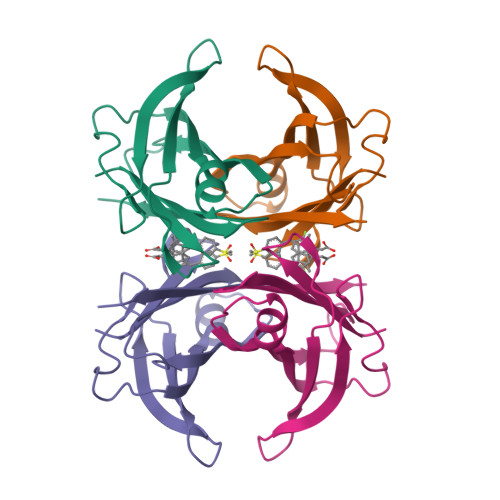
Crystal structure of wild-type human transthyretin in complex with sulindacTo be published
Crystal structure of wild-type human transthyretin in complex with sulindacTo be published
Crystal structure of wild-type human transthyretin in complex with sulindacLima, L.M.T.R., Polikarpov, I. To be published
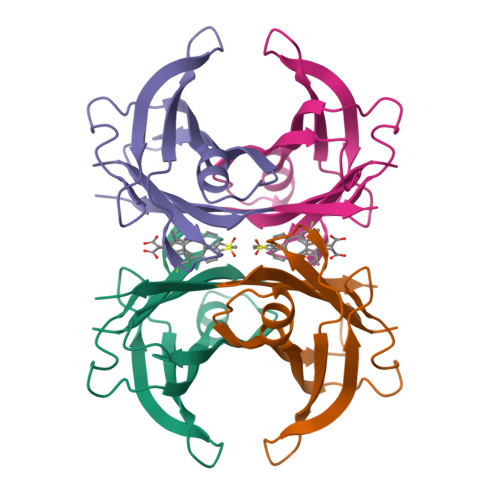
AKR1C2 complex with sulindacYosaatmadja, Y., Flanagan, J.U., Squire, C.J. To be published
Crystal structure of human AKR1B10 complexed with NADP+ and sulindacCousido-Siah, A., Ruiz, F.X., Mitschler, A., Crespo, I., Porte, S., Pares, X., Farres, J., Podjarny, A. (2015) Chem Biol Interact 234: 290-296
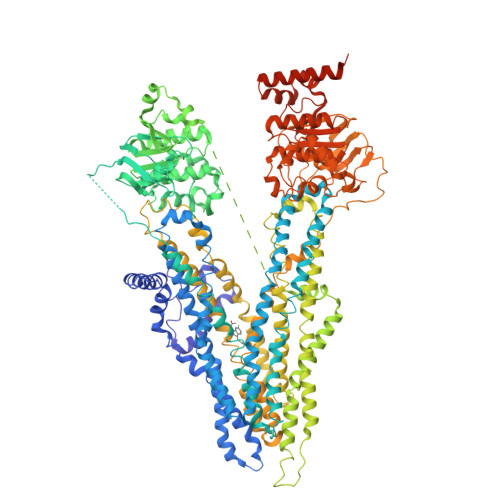
cryo-EM structure of sulindac-bound hMRP4(2023) Commun Biol 6: 549-549
1 to 10 of 10 Structures Page 1 of 1 Sort by |