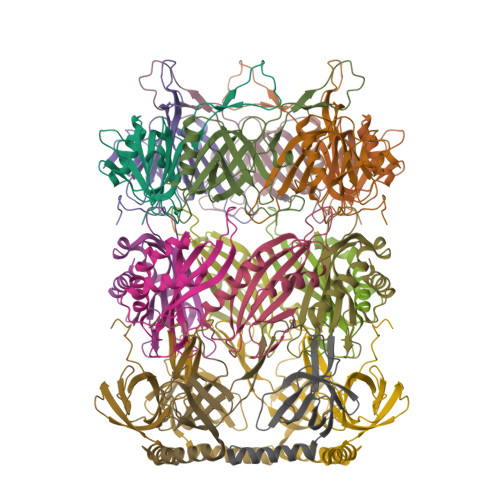
Structure Determination MethodologyScientific Name of Source OrganismRefinement Resolution (Å)Enzyme Classification NameMembrane Protein Annotation | Yin, Z., Ebright, R.H. (2022) Proc Natl Acad Sci U S A 119: e2205278119-e2205278119 | Released | 2022-09-28 | | Method | X-RAY DIFFRACTION 1.46 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CL, EDO, GOL, PEG, PO4, ZN |
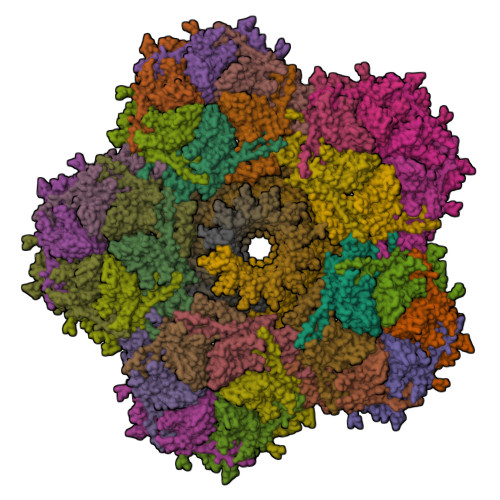
Yin, Z., Ebright, R.H. (2022) Proc Natl Acad Sci U S A 119: e2205278119-e2205278119 | Released | 2022-09-28 | | Method | X-RAY DIFFRACTION 1.97 Å | | Organisms | | | Macromolecule | | | Unique Ligands | CL, EDO, GOL, PEG, ZN |
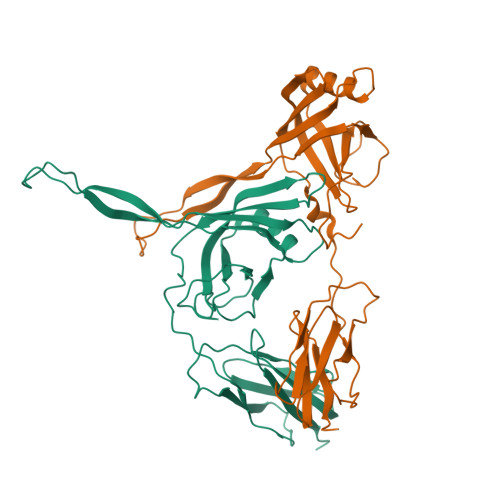
Yin, Z., Ebright, R.H. (2022) Proc Natl Acad Sci U S A 119: e2205278119-e2205278119 | Released | 2022-09-28 | | Method | X-RAY DIFFRACTION 2.177 Å | | Organisms | | | Macromolecule | | | Unique Ligands | EDO, ZN |
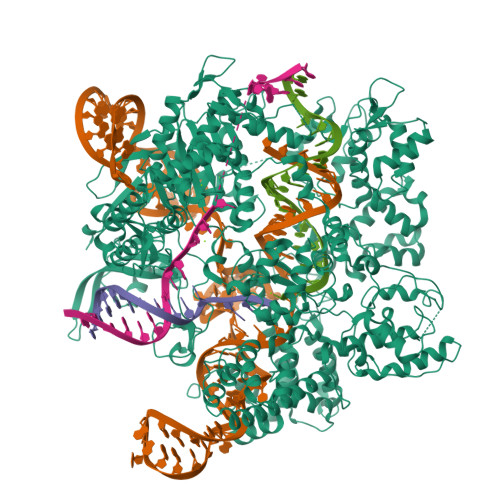
Yin, Z., Ebright, R.H. (2022) Proc Natl Acad Sci U S A 119: e2205278119-e2205278119 | Released | 2022-09-28 | | Method | ELECTRON MICROSCOPY 3.13 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 Unique nucleic acid chains: 3 | | Unique Ligands | MG, ZN |
Yin, Z., Ebright, R.H. (2022) Proc Natl Acad Sci U S A 119: e2205278119-e2205278119 | Released | 2022-09-28 | | Method | ELECTRON MICROSCOPY 3.36 Å | | Organisms | | | Macromolecule | Unique protein chains: 7 Unique nucleic acid chains: 3 | | Unique Ligands | MG, ZN |
Florez Ariza, A., Wee, L., Tong, A., Canari, C., Grob, P., Nogales, E., Bustamante, C. (2023) Cell 186: 1244-1262.e34 | Released | 2023-03-29 | | Method | ELECTRON MICROSCOPY 3.9 Å | | Organisms | | | Macromolecule | Unique protein chains: 4 Unique nucleic acid chains: 3 | | Unique Ligands | MG, ZN |
Florez Ariza, A., Wee, L., Tong, A., Canari, C., Grob, P., Nogales, E., Bustamante, C. (2023) Cell 186: 1244-1262.e34 | Released | 2023-03-29 | | Method | ELECTRON MICROSCOPY 7.3 Å | | Organisms | | | Macromolecule | Unique protein chains: 3 Unique nucleic acid chains: 3 | | Unique Ligands | MG, ZN |
Florez Ariza, A., Wee, L., Tong, A., Canari, C., Grob, P., Nogales, E., Bustamante, C. (2023) Cell 186: 1244-1262.e34 | Released | 2023-03-29 | | Method | ELECTRON MICROSCOPY 3.8 Å | | Organisms | | | Macromolecule | Unique protein chains: 51 Unique nucleic acid chains: 5 |
Gao, B., Feng, Y. (2023) Proc Natl Acad Sci U S A 120: e2217493120-e2217493120 | Released | 2022-12-21 | | Method | ELECTRON MICROSCOPY 2.78 Å | | Organisms | | | Macromolecule | | | Unique Ligands | AGS, MG |
Wang, J.W., Wang, C. (2024) Structure 32: 35 | Released | 2023-10-18 | | Method | ELECTRON MICROSCOPY 3.1 Å | | Organisms | | | Macromolecule | |
Wang, J.W., Wang, C. (2024) Structure 32: 35 | Released | 2023-10-18 | | Method | ELECTRON MICROSCOPY 2.95 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | SF4 |
Wang, J.W., Wang, C. (2024) Structure 32: 35 | Released | 2023-10-18 | | Method | ELECTRON MICROSCOPY 3 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | SF4 |
Wang, J. (2024) Structure 32: 35 | Released | 2023-10-18 | | Method | ELECTRON MICROSCOPY 3.86 Å | | Organisms | | | Macromolecule | |
Xiao, H., Tan, L., Cheng, L.P., Liu, H.R. (2023) PLoS Biol 21: e3002441-e3002441 | Released | 2023-11-15 | | Method | ELECTRON MICROSCOPY 3.44 Å | | Organisms | | | Macromolecule | Unique protein chains: 6 | | Unique Ligands | SF4 |
Wang, J.W. (2024) Structure 32: 35 | Released | 2023-10-18 | | Method | ELECTRON MICROSCOPY 3.8 Å | | Organisms | | | Macromolecule | |
|