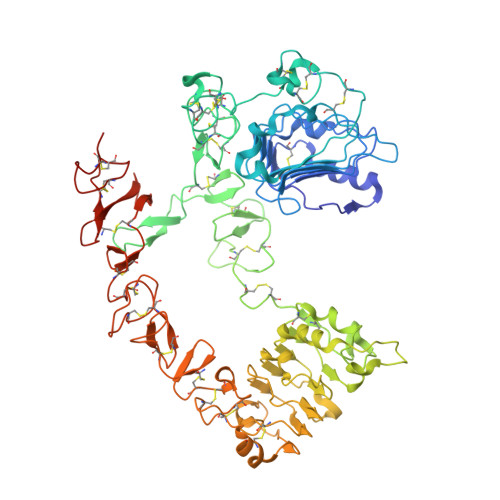
EGF activates its receptor by removing interactions that auto-inhibit ectodomain dimerization
Ferguson, K.M., Berger, M.B., Mendrola, J.M., Cho, H., Leahy, D.J., Lemmon, M.A.(2003) Mol Cell 11: 507-517
- PubMed: 12620237
- DOI: https://doi.org/10.1016/s1097-2765(03)00047-9
- Primary Citation of Related Structures:
1NQL - PubMed Abstract:
Epidermal growth factor (EGF) receptor is the prototype of the ErbB (HER) family receptor tyrosine kinases (RTKs), which regulate cell growth and differentiation and are implicated in many human cancers. EGF activates its receptor by inducing dimerization of the 621 amino acid EGF receptor extracellular region. We describe the 2.8 A resolution crystal structure of this entire extracellular region (sEGFR) in an unactivated state. The structure reveals an autoinhibited configuration, where the dimerization interface recently identified in activated sEGFR structures is completely occluded by intramolecular interactions. To activate the receptor, EGF binding must promote a large domain rearrangement that exposes this dimerization interface. This contrasts starkly with other RTK activation mechanisms and suggests new approaches for designing ErbB receptor antagonists.
Organizational Affiliation:
Department of Biochemistry and Biophysics, University of Pennsylvania School of Medicine, Philadelphia, PA 19104, USA. fertuso2@mail.med.upenn.edu