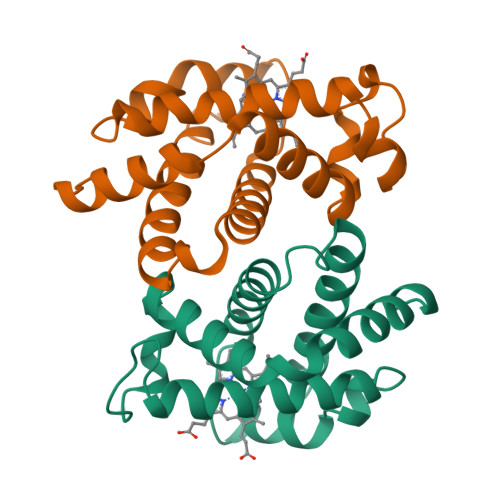
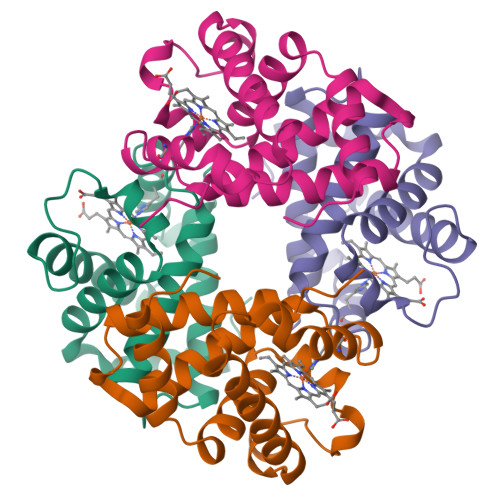
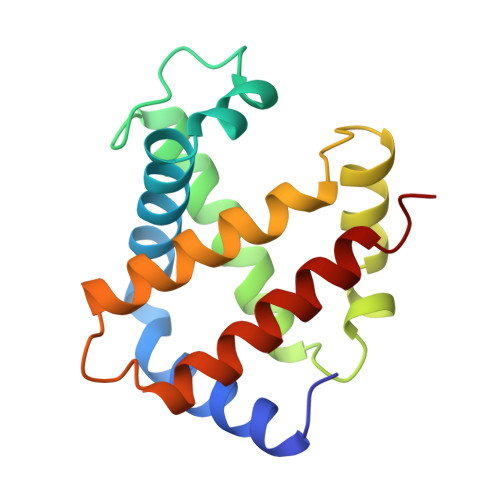
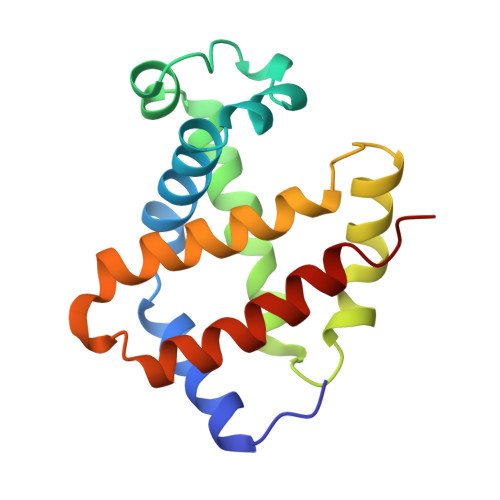
The crystal structures of trout Hb I in the deoxy and carbonmonoxy forms.
Tame, J.R., Wilson, J.C., Weber, R.E.(1996) J Mol Biology 259: 749-760
- PubMed: 8683580
- DOI: https://doi.org/10.1006/jmbi.1996.0355
- Primary Citation of Related Structures:
1OUT, 1OUU - PubMed Abstract:
We have determined the X-ray crystallographic structure of trout Hb I in both the deoxy and carbonmonoxy forms to resolution limits of 2.3 angstroms and 2.5 angstroms, respectively. The overall fold of the molecule is highly similar to that of human HbA despite the low level of sequence identity between these proteins. Trout Hb I is unusual in displaying almost no pH dependence of oxygen binding affinity, and (at most) very weak interactions with heterotropic effector ligands such as organic phosphates. Comparison of the two quaternary states of the protein indicates how such effects are minimised and how the low-affinity T state of the protein is stabilised in the absence of heterotropic interactions.
- Department of Chemistry, University of York, Helsington, UK.
Organizational Affiliation: