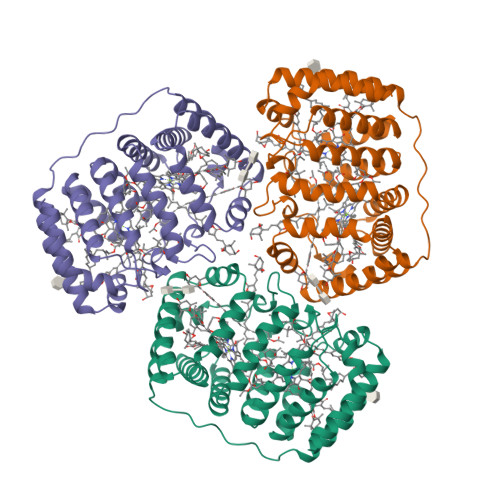
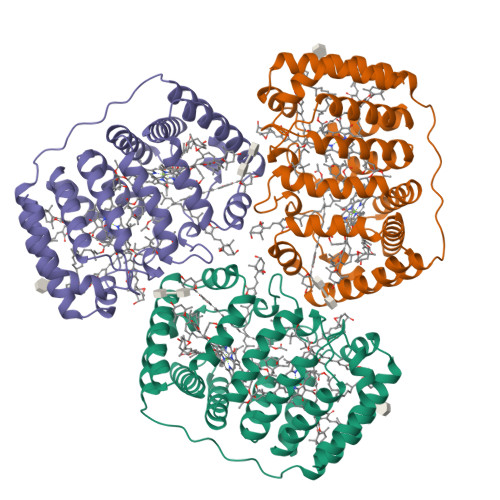
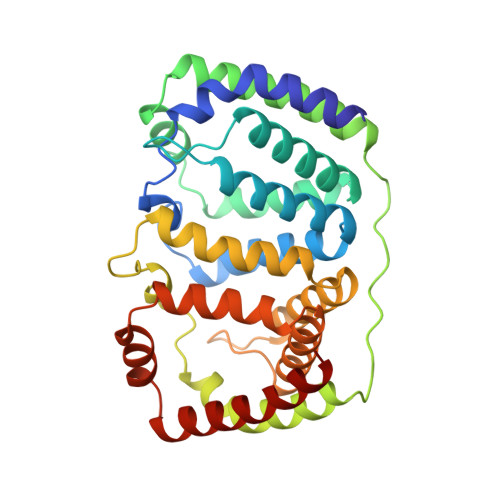
Structural basis of light harvesting by carotenoids: peridinin-chlorophyll-protein from Amphidinium carterae.
Hofmann, E., Wrench, P.M., Sharples, F.P., Hiller, R.G., Welte, W., Diederichs, K.(1996) Science 272: 1788-1791
- PubMed: 8650577
- DOI: https://doi.org/10.1126/science.272.5269.1788
- Primary Citation of Related Structures:
1PPR - PubMed Abstract:
Peridinin-chlorophyll-protein, a water-soluble light-harvesting complex that has a blue-green absorbing carotenoid as its main pigment, is present in most photosynthetic dinoflagellates. Its high-resolution (2.0 angstrom) x-ray structure reveals a noncrystallographic trimer in which each polypeptide contains an unusual jellyroll fold of the alpha-helical amino- and carboxyl-terminal domains. These domains constitute a scaffold with pseudo-twofold symmetry surrounding a hydrophobic cavity filled by two lipid, eight peridinin, and two chlorophyll a molecules. The structural basis for efficient excitonic energy transfer from peridinin to chlorophyll is found in the clustering of peridinins around the chlorophylls at van der Waals distances.
Organizational Affiliation:
Fakultät für Biologie, Universität Konstanz, Germany.