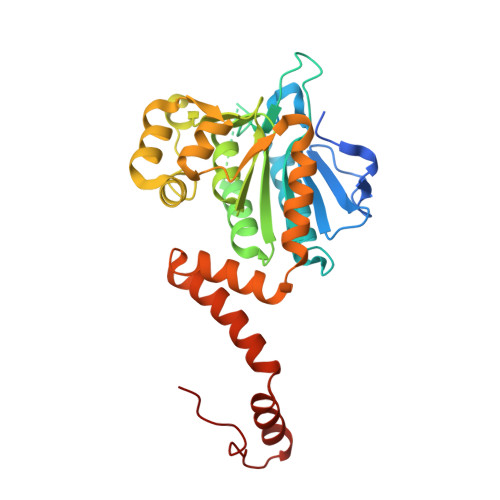
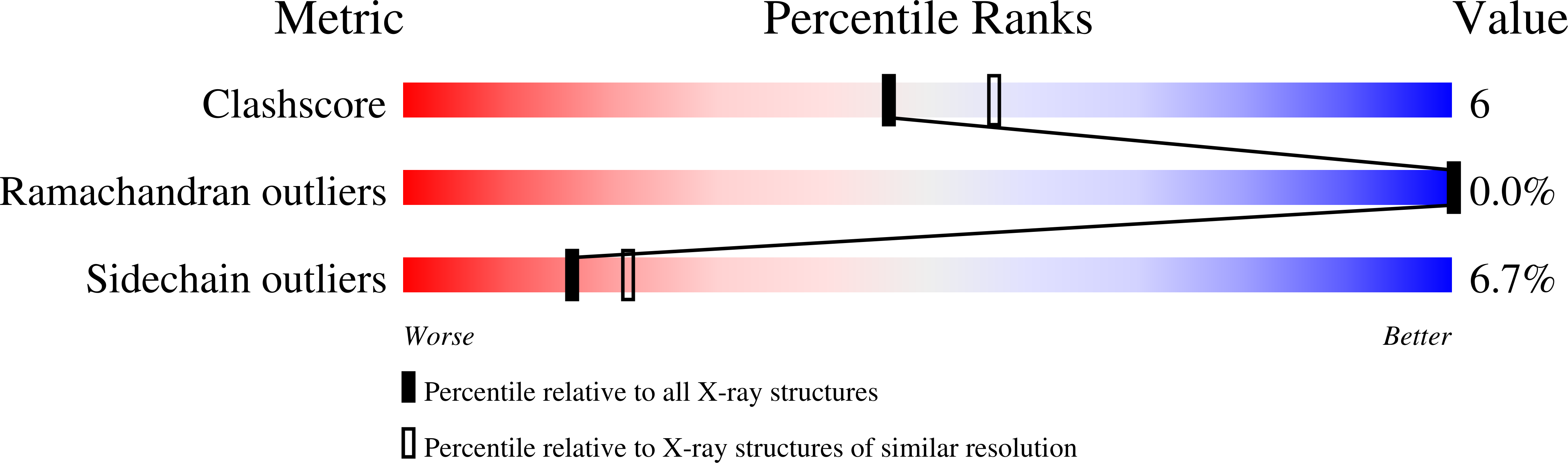Crystal structure of Mycobacterium tuberculosis MenB, a key enzyme in vitamin K2 biosynthesis.
Truglio, J.J., Theis, K., Feng, Y., Gajda, R., Machutta, C., Tonge, P.J., Kisker, C.(2003) J Biol Chem 278: 42352-42360
- PubMed: 12909628
- DOI: https://doi.org/10.1074/jbc.M307399200
- Primary Citation of Related Structures:
1Q51, 1Q52 - PubMed Abstract:
Bacterial enzymes of the menaquinone (Vitamin K2) pathway are potential drug targets because they lack human homologs. MenB, 1,4-dihydroxy-2-naphthoyl-CoA synthase, the fourth enzyme in the biosynthetic pathway leading from chorismate to menaquinone, catalyzes the conversion of O-succinylbenzoyl-CoA (OSB-CoA) to 1,4-dihydroxy-2-naphthoyl-CoA (DHNA-CoA). Based on our interest in developing novel tuberculosis chemotherapeutics, we have solved the structures of MenB from Mycobacterium tuberculosis and its complex with acetoacetyl-coenzyme A at 1.8 and 2.3 A resolution, respectively. Like other members of the crotonase superfamily, MenB folds as an (alpha3)2 hexamer, but its fold is distinct in that the C terminus crosses the trimer-trimer interface, forming a flexible part of the active site within the opposing trimer. The highly conserved active site of MenB contains a deep pocket lined by Asp-192, Tyr-287, and hydrophobic residues. Mutagenesis shows that Asp-192 and Tyr-287 are essential for enzymatic catalysis. We postulate a catalytic mechanism in which MenB enables proton transfer within the substrate to yield an oxyanion as the initial step in catalysis. Knowledge of the active site geometry and characterization of the catalytic mechanism of MenB will aid in identifying new inhibitors for this potential drug target.
Organizational Affiliation:
Department of Pharmacological Sciences, Center for Structural Biology, State University of New York at Stony Brook, NY 11794-5115, USA.