Structure and catalytic properties of an engineered heterodimer of enolase composed of one active and one inactive subunit
Sims, P.A., Menefee, A.L., Larsen, T.M., Mansoorabadi, S.O., Reed, G.H.(2006) J Mol Biology 355: 422-431
- PubMed: 16309698
- DOI: https://doi.org/10.1016/j.jmb.2005.10.050
- Primary Citation of Related Structures:
2AL1, 2AL2 - PubMed Abstract:
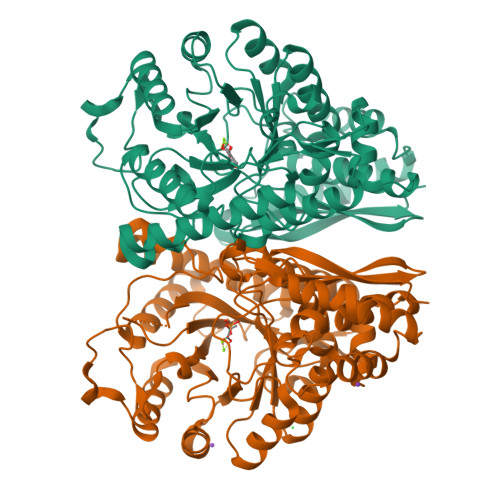
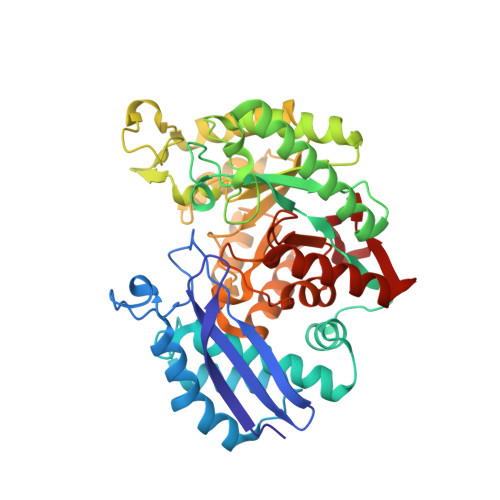
Enolase is a dimeric enzyme that catalyzes the interconversion of 2-phospho-D-glycerate and phosphoenolpyruvate. This reversible dehydration is effected by general acid-base catalysis that involves, principally, Lys345 and Glu211 (numbering system of enolase 1 from yeast). The crystal structure of the inactive E211Q enolase shows that the protein is properly folded. However, K345 variants have, thus far, failed to crystallize. This problem was solved by crystallization of an engineered heterodimer of enolase. The heterodimer was composed of an inactive subunit that has a K345A mutation and an active subunit that has N80D and N126D surface mutations to facilitate ion-exchange chromatographic separation of the three dimeric species. The structure of this heterodimeric variant, in complex with substrate/product, was obtained at 1.85 A resolution. The structure was compared to a new structure of wild-type enolase obtained from crystals belonging to the same space group. Asymmetric dimers having one subunit exhibiting two of the three active site loops in an open conformation and the other in a conformation having all three loops closed appear in both structures. The K345A subunit of the heterodimer is in the loop-closed conformation; its Calpha carbon atoms closely match those of the corresponding subunit of wild-type enolase (root-mean-squared deviation of 0.23 A). The kcat and kcat/Km values of the heterodimer are approximately half those of the N80D/N126D homodimer, which suggests that the subunits in solution are kinetically independent. A comparison of enolase structures obtained from crystals belonging to different space groups suggests that asymmetric dimers can be a consequence of the asymmetric positioning of the subunits within the crystal lattice.
Organizational Affiliation:
Department of Biochemistry, University of Wisconsin-Madison, Madison, WI, 53726, USA.