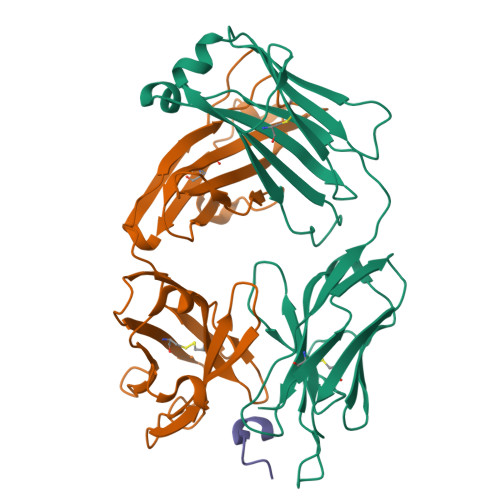
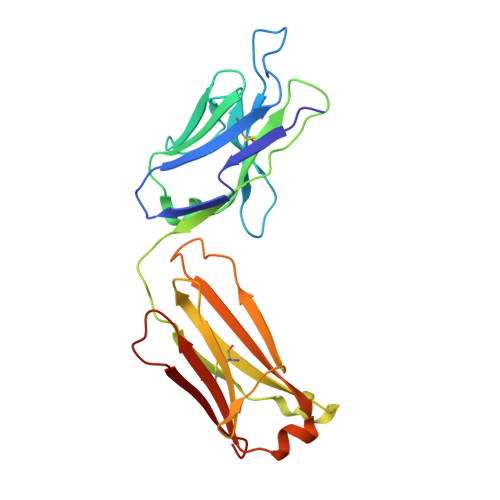
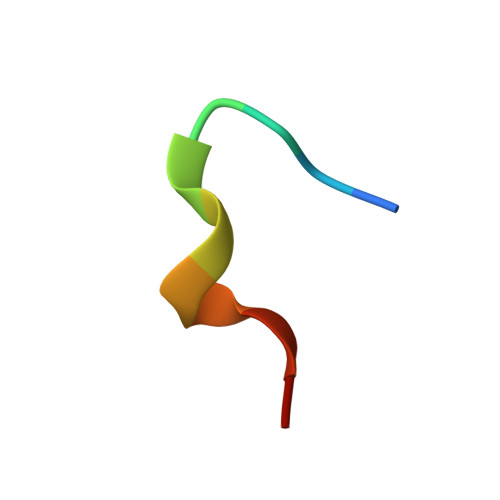Broadly neutralizing anti-hepatitis B virus antibody reveals a complementarity determining region H3 lid-opening mechanism
Chi, S.-W., Maeng, C.-Y., Kim, S.J., Oh, M.S., Ryu, C.J., Kim, S.-J., Han, K.-H., Hong, H.J., Ryu, S.-E.(2007) Proc Natl Acad Sci U S A 104: 9230-9235
- PubMed: 17517649
- DOI: https://doi.org/10.1073/pnas.0701279104
- Primary Citation of Related Structures:
2EH7, 2EH8 - PubMed Abstract:
The humanized monoclonal antibody HzKR127 recognizes the preS1 domain of the human hepatitis B virus surface proteins with a broadly neutralizing activity in vivo. We present the crystal structures of HzKR127 Fab and its complex with a major epitope peptide. In the complex structure, the bound peptide forms a type IV beta-turn followed by 3(10) helical turn, the looped-out conformation of which provides a structural basis for broad neutralization. Upon peptide binding, the antibody undergoes a dramatic complementarity determining region H3 lid opening. To understand the structural implication of the virus neutralization, we carried out comprehensive alanine-scanning mutagenesis of all complementarity determining region residues in HzKR127 Fab. The functional mapping of the antigen-combining site demonstrates the specific roles of major binding determinants in antigen binding, contributing to the rational design for maximal humanization and affinity maturation of the antibody.
Organizational Affiliation:
Center for Cellular Switch Protein Structure, Molecular Cancer Research Center, Korea Research Institute of Bioscience and Biotechnology, Daejeon 305-333, Korea.