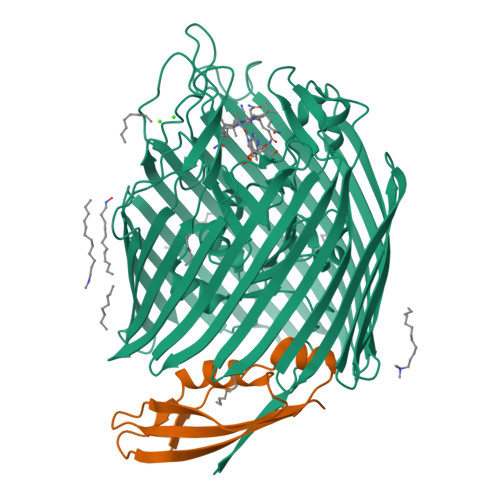
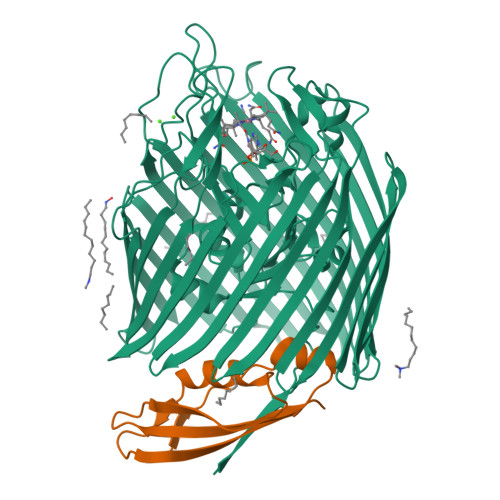
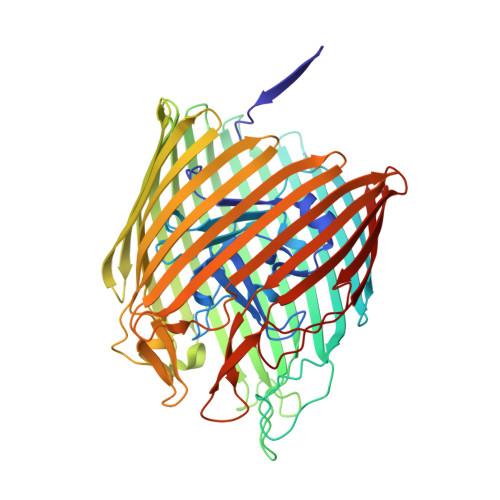
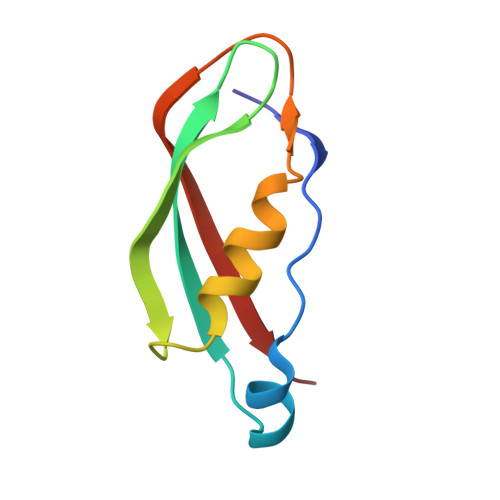
Outer membrane active transport: structure of the BtuB:TonB complex
Shultis, D.D., Purdy, M.D., Banchs, C.N., Wiener, M.C.(2006) Science 312: 1396-1399
- PubMed: 16741124
- DOI: https://doi.org/10.1126/science.1127694
- Primary Citation of Related Structures:
2GSK - PubMed Abstract:
In Gram-negative bacteria, the import of essential micronutrients across the outer membrane requires a transporter, an electrochemical gradient of protons across the inner membrane, and an inner membrane protein complex (ExbB, ExbD, TonB) that couples the proton-motive force to the outer membrane transporter. The inner membrane protein TonB binds directly to a conserved region, called the Ton-box, of the transporter. We solved the structure of the cobalamin transporter BtuB in complex with the C-terminal domain of TonB. In contrast to its conformations in the absence of TonB, the Ton-box forms a beta strand that is recruited to the existing beta sheet of TonB, which is consistent with a mechanical pulling model of transport.
Organizational Affiliation:
Department of Molecular Physiology and Biological Physics, University of Virginia, Charlottesville, VA 22908, USA.