The Structures of l-Rhamnose Isomerase from Pseudomonas stutzeri in Complexes with l-Rhamnose and d-Allose Provide Insights into Broad Substrate Specificity
Yoshida, H., Yamada, M., Ohyama, Y., Takada, G., Izumori, K., Kamitori, S.(2007) J Mol Biology 365: 1505-1516
- PubMed: 17141803
- DOI: https://doi.org/10.1016/j.jmb.2006.11.004
- Primary Citation of Related Structures:
2HCV, 2I56, 2I57 - PubMed Abstract:
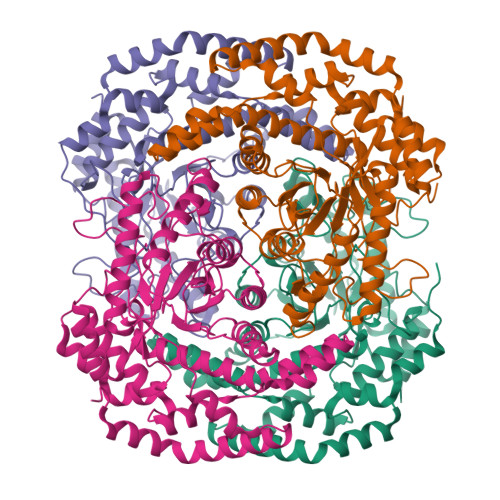
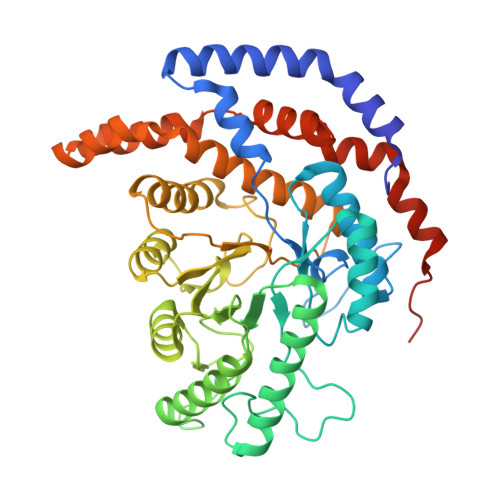
Pseudomonas stutzeri L-rhamnose isomerase (P. stutzeri L-RhI) can efficiently catalyze the isomerization between various aldoses and ketoses, showing a broad substrate specificity compared to L-RhI from Escherichia coli (E. coli L-RhI). To understand the relationship between structure and substrate specificity, the crystal structures of P. stutzeri L-RhI alone and in complexes with L-rhamnose and D-allose which has different configurations of C4 and C5 from L-rhamnose, were determined at a resolution of 2.0 A, 1.97 A, and 1.97 A, respectively. P. stutzeri L-RhI has a large domain with a (beta/alpha)(8) barrel fold and an additional small domain composed of seven alpha-helices, forming a homo tetramer, as found in E. coli L-RhI and D-xylose isomerases (D-XIs) from various microorganisms. The beta1-alpha1 loop (Gly60-Arg76) of P. stutzeri L-RhI is involved in the substrate binding of a neighbouring molecule, as found in D-XIs, while in E. coli L-RhI, the corresponding beta1-alpha1 loop is extended (Asp52-Arg78) and covers the substrate-binding site of the same molecule. The complex structures of P. stutzeri L-RhI with L-rhamnose and D-allose show that both substrates are nicely fitted to the substrate-binding site. The part of the substrate-binding site interacting with the substrate at the 1, 2, and 3 positions is equivalent to E. coli L-RhI, and the other part interacting with the 4, 5, and 6 positions is similar to D-XI. In E. coli L-RhI, the beta1-alpha1 loop creates an unique hydrophobic pocket at the the 4, 5, and 6 positions, leading to the strictly recognition of L-rhamnose as the most suitable substrate, while in P. stutzeri L-RhI, there is no corresponding hydrophobic pocket where Phe66 from a neighbouring molecule merely forms hydrophobic interactions with the substrate, leading to the loose substrate recognition at the 4, 5, and 6 positions.
Organizational Affiliation:
Molecular Structure Research Group, Information Technology Center and Faculty of Medicine, Kagawa University, 1750-1 Ikenobe, Miki-cho, Kita-gun, Kagawa 761-0793, Japan.