Functional Significance of Four Successive Glycine Residues in the Pyrophosphate Binding Loop of Fungal 6-Oxopurine Phosphoribosyltransferases.
Moynie, L., Giraud, M.F., Breton, A., Boissier, F., Daignan-Fornier, B., Dautant, A.(2012) Protein Sci 21: 1185
- PubMed: 22610485
- DOI: https://doi.org/10.1002/pro.2098
- Primary Citation of Related Structures:
2JKY, 2JKZ, 2XBU - PubMed Abstract:
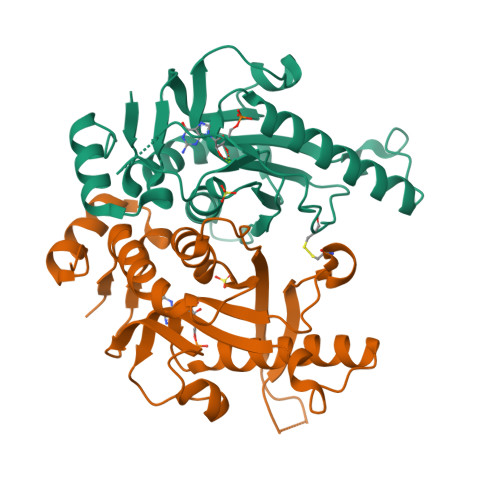
Hypoxanthine-guanine phosphoribosyltransferase (HGPRT) is a key enzyme of the purine recycling pathway that catalyzes the conversion of 5-phospho-ribosyl-α-1-pyrophosphate and guanine or hypoxanthine to guanosine monophosphate (GMP) or inosine monophosphate (IMP), respectively, and pyrophosphate (PPi). We report the first crystal structure of a fungal 6-oxopurine phosphoribosyltransferase, the Saccharomyces cerevisiae HGPRT (Sc-HGPRT) in complex with GMP. The crystal structures of full length protein with (WT1) or without (WT2) sulfate that mimics the phosphate group in the PPi binding site were solved by molecular replacement using the structure of a truncated version (Δ7) solved beforehand by multiwavelength anomalous diffusion. Sc-HGPRT is a dimer and adopts the overall structure of class I phosphoribosyltransferases (PRTs) with a smaller hood domain and a short two-stranded parallel β-sheet linking the N- to the C-terminal end. The catalytic loops in WT1 and WT2 are in an open form while in Δ7, due to an inter-subunit disulfide bridge, the catalytic loop is in either an open or closed form. The closure is concomitant with a peptide plane flipping in the PPi binding loop. Moreover, owing the flexibility of a GGGG motif conserved in fungi, all the peptide bonds of the phosphate binding loop are in trans conformation whereas in nonfungal 6-oxopurine PRTs, one cis-peptide bond is required for phosphate binding. Mutations affecting the enzyme activity or the previously characterized feedback inhibition by GMP are located at the nucleotide binding site and the dimer interface.
Organizational Affiliation:
IBGC, Université de Bordeaux, UMR 5095, F-33000 Bordeaux, France.