Structures of the dI(2)dIII(1) Complex of Proton-Translocating Transhydrogenase with Bound, Inactive Analogues of NADH and NADPH Reveal Active Site Geometries
Bhakta, T., Whitehead, S.J., Snaith, J.S., Dafforn, T.R., Wilkie, J., Rajesh, S., White, S.A., Jackson, J.B.(2007) Biochemistry 46: 3304-3318
- PubMed: 17323922
- DOI: https://doi.org/10.1021/bi061843r
- Primary Citation of Related Structures:
2OO5, 2OOR - PubMed Abstract:
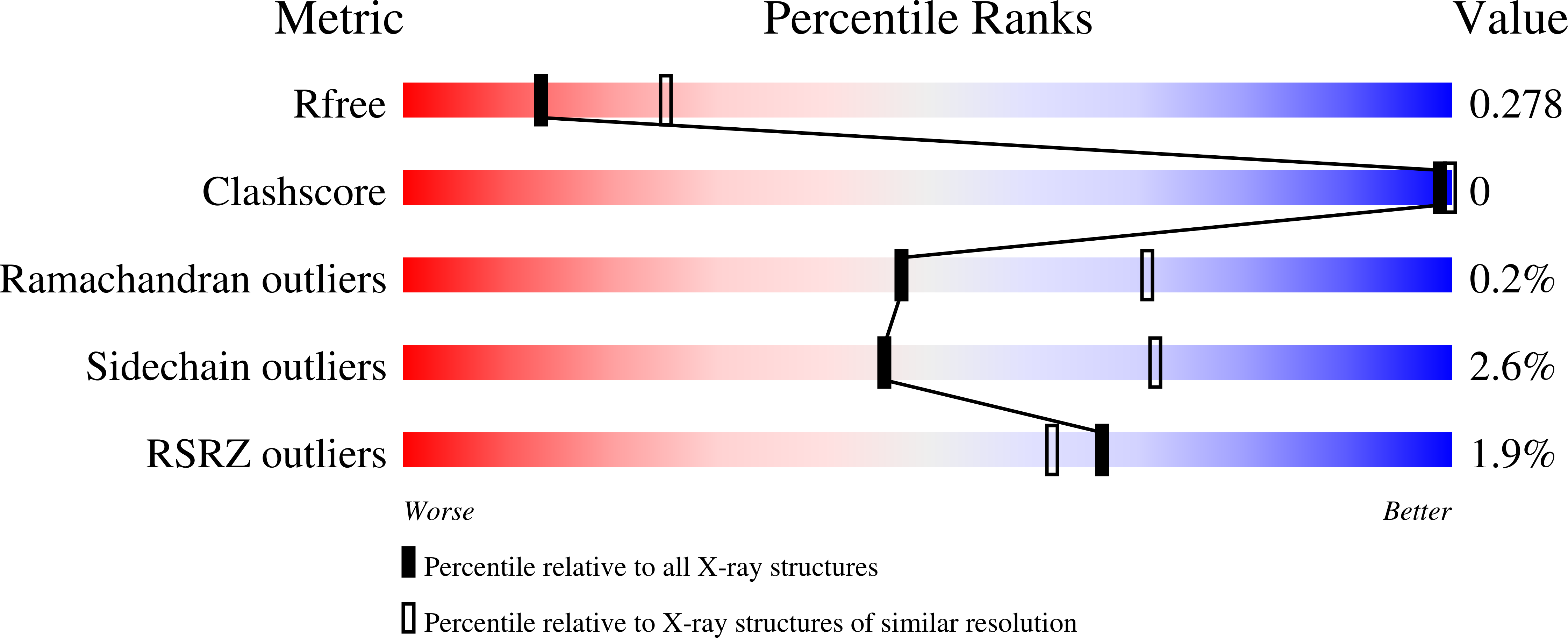
Transhydrogenase couples the redox reaction between NADH and NADP+ to proton translocation across a membrane. The enzyme comprises three components; dI binds NAD(H), dIII binds NADP(H), and dII spans the membrane. The 1,4,5,6-tetrahydro analogue of NADH (designated H2NADH) bound to isolated dI from Rhodospirillum rubrum transhydrogenase with similar affinity to the physiological nucleotide. Binding of either NADH or H2NADH led to closure of the dI mobile loop. The 1,4,5,6-tetrahydro analogue of NADPH (H2NADPH) bound very tightly to isolated R. rubrum dIII, but the rate constant for dissociation was greater than that for NADPH. The replacement of NADP+ on dIII either with H2NADPH or with NADPH caused a similar set of chemical shift alterations, signifying an equivalent conformational change. Despite similar binding properties to the natural nucleotides, neither H2NADH nor H2NADPH could serve as a hydride donor in transhydrogenation reactions. Mixtures of dI and dIII form dI2dIII1 complexes. The nucleotide charge distribution of complexes loaded either with H2NADH and NADP+ or with NAD+ and H2NADPH should more closely mimic the ground states for forward and reverse hydride transfer, respectively, than previously studied dead-end species. Crystal structures of such complexes at 2.6 and 2.3 A resolution are described. A transition state for hydride transfer between dihydronicotinamide and nicotinamide derivatives determined in ab initio quantum mechanical calculations resembles the organization of nucleotides in the transhydrogenase active site in the crystal structure. Molecular dynamics simulations of the enzyme indicate that the (dihydro)nicotinamide rings remain close to a ground state for hydride transfer throughout a 1.4 ns trajectory.
Organizational Affiliation:
School of Biosciences, University of Birmingham, Edgbaston, UK.