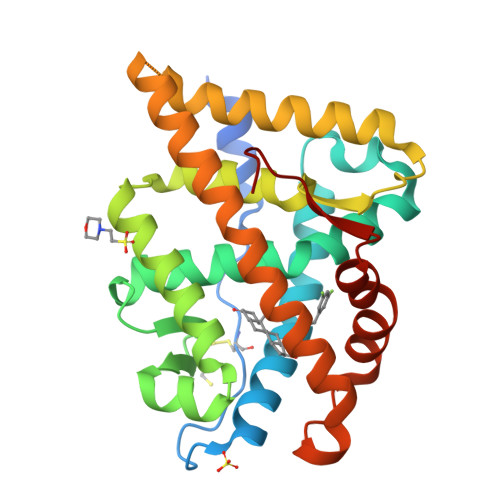
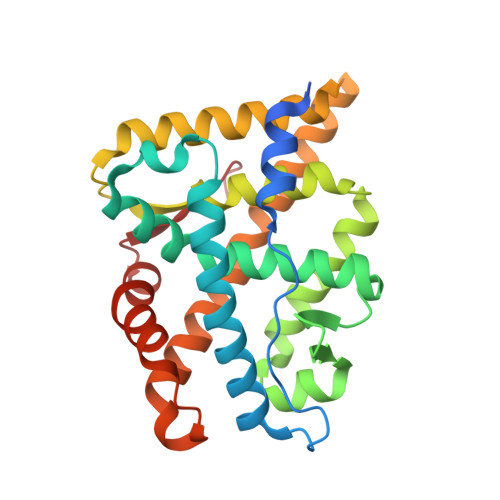
Structural Characterization of the Human Androgen Receptor Ligand-binding Domain Complexed with EM5744, a Rationally Designed Steroidal Ligand Bearing a Bulky Chain Directed toward Helix 12.
Cantin, L., Faucher, F., Couture, J.F., de Jesus-Tran, K.P., Legrand, P., Ciobanu, L.C., Frechette, Y., Labrecque, R., Singh, S.M., Labrie, F., Breton, R.(2007) J Biological Chem 282: 30910-30919
- PubMed: 17711855
- DOI: https://doi.org/10.1074/jbc.M705524200
- Primary Citation of Related Structures:
2PNU - PubMed Abstract:
Antiandrogens are commonly used to treat androgen-dependent disorders. The currently used drugs unfortunately possess very weak affinity for the human AR (hAR), thus indicating the need to develop new high-affinity steroidal antiandrogens. Our compounds are specially designed to impede repositioning of the mobile carboxyl-terminal helix 12, which blocks the ligand-dependent transactivation function (AF-2) located in the AR ligand-binding domain (ARLBD). Using crystal structures of the hARLBD, we first found that H12 could be directly reached from the ligand-binding pocket (LBP) by a chain positioned on the C18 atom of an androgen steroid nucleus. A set of 5alpha-dihydrotestosterone-derived molecules bearing various C18 chains were thus synthesized and tested for their capacity to bind hAR and act as antagonists. Although most of those having very high affinity for hAR were agonists, several very potent antagonists were obtained, confirming the structural importance of the C18 chain. To understand the role of the C18 chain in their agonistic/antagonistic properties, the structure of the hARLBD complexed with one of these agonists, EM5744, was determined at a 1.65-A resolution. We have identified new interactions involving Gln(738), Met(742), and His(874) that explain both the high affinity of this compound and the inability of its bulky chain to prevent the repositioning of H12. This structural information will be helpful to refine the structure of the chains placed on the C18 atom to obtain efficient H12-directed steroidal antiandrogens.
Organizational Affiliation:
Oncology and Molecular Endocrinology Research Center, Laval University Medical Center (CHUL) and Laval University, Québec G1V 4G2, Canada.