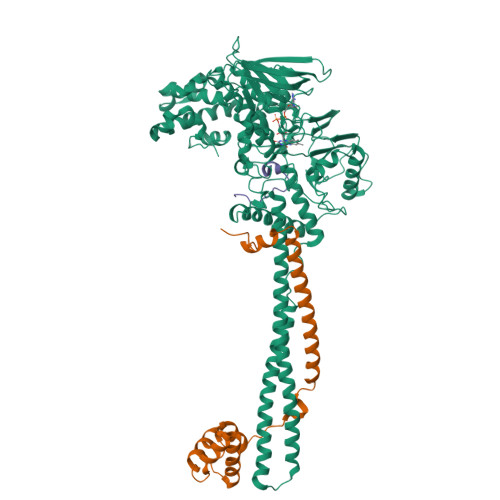
Structural Basis of Lsd1-Corest Selectivity in Histone H3 Recognition.
Forneris, F., Binda, C., Adamo, A., Battaglioli, E., Mattevi, A.(2007) J Biological Chem 282: 20070
- PubMed: 17537733
- DOI: https://doi.org/10.1074/jbc.C700100200
- Primary Citation of Related Structures:
2V1D - PubMed Abstract:
Histone demethylase LSD1 regulates transcription by demethylating Lys(4) of histone H3. The crystal structure of the enzyme in complex with CoREST and a substrate-like peptide inhibitor highlights an intricate network of interactions and a folded conformation of the bound peptide. The core of the peptide structure is formed by Arg(2), Gln(5), and Ser(10), which are engaged in specific intramolecular H-bonds. Several charged side chains on the surface of the substrate-binding pocket establish electrostatic interactions with the peptide. The three-dimensional structure predicts that methylated Lys(4) binds in a solvent inaccessible position in front of the flavin cofactor. This geometry is fully consistent with the demethylation reaction being catalyzed through a flavin-mediated oxidation of the substrate amino-methyl group. These features dictate the exquisite substrate specificity of LSD1 and provide a structural framework to explain the fine tuning of its catalytic activity and the active role of CoREST in substrate recognition.
Organizational Affiliation:
Dipartimento di Genetica e Microbiologia, Università di Pavia, Via Ferrata 1, 27100 Pavia, Italy.