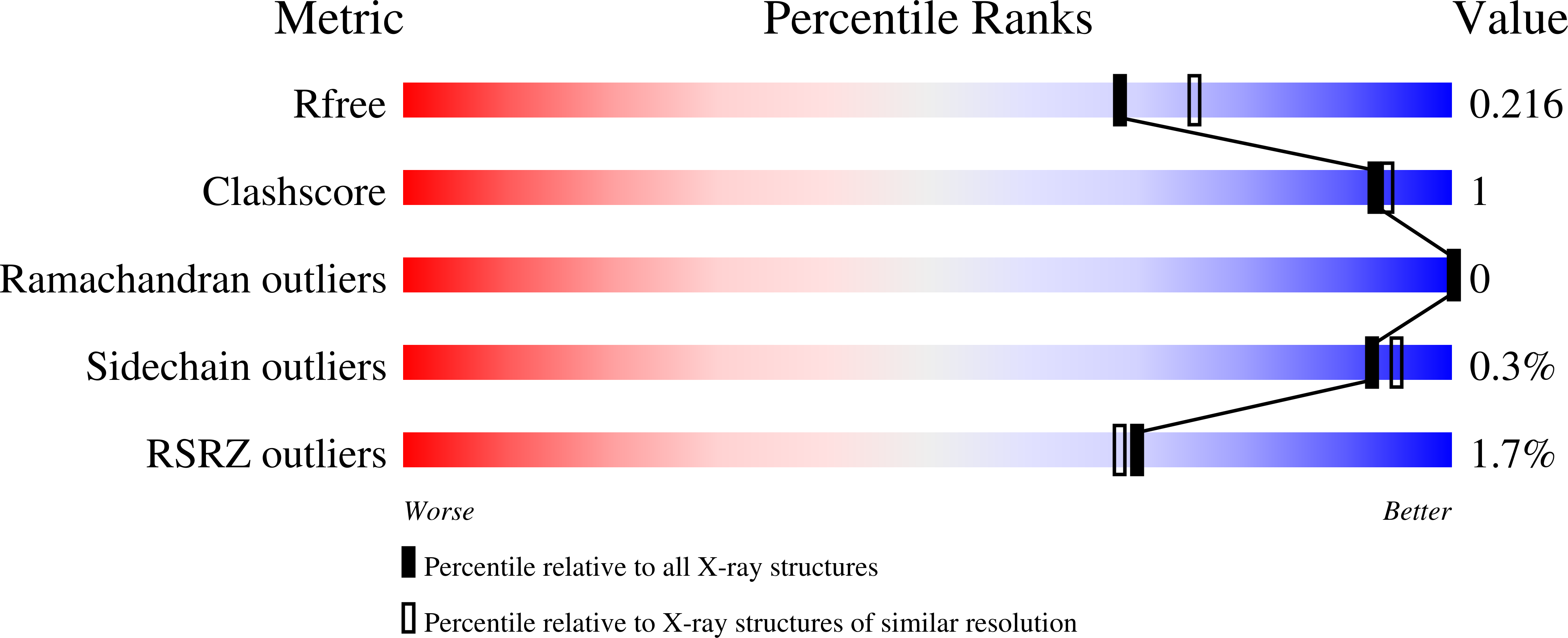
Active site dynamics in the zinc-dependent medium chain alcohol dehydrogenase superfamily.
Baker, P.J., Britton, K.L., Fisher, M., Esclapez, J., Pire, C., Bonete, M.J., Ferrer, J., Rice, D.W.(2009) Proc Natl Acad Sci U S A 106: 779-784
- PubMed: 19131516
- DOI: https://doi.org/10.1073/pnas.0807529106
- Primary Citation of Related Structures:
2VWG, 2VWH, 2VWP, 2VWQ - PubMed Abstract:
Despite being the subject of intensive investigations, many aspects of the mechanism of the zinc-dependent medium chain alcohol dehydrogenase (MDR) superfamily remain contentious. We have determined the high-resolution structures of a series of binary and ternary complexes of glucose dehydrogenase, an MDR enzyme from Haloferax mediterranei. In stark contrast to the textbook MDR mechanism in which the zinc ion is proposed to remain stationary and attached to a common set of protein ligands, analysis of these structures reveals that in each complex, there are dramatic differences in the nature of the zinc ligation. These changes arise as a direct consequence of linked movements of the zinc ion, a zinc-bound bound water molecule, and the substrate during progression through the reaction. These results provide evidence for the molecular basis of proton traffic during catalysis, a structural explanation for pentacoordinate zinc ion intermediates, a unifying view for the observed patterns of metal ligation in the MDR family, and highlight the importance of dynamic fluctuations at the metal center in changing the electrostatic potential in the active site, thereby influencing the proton traffic and hydride transfer events.
Organizational Affiliation:
The Krebs Institute, Department of Molecular Biology and Biotechnology, University of Sheffield, Western Bank, Sheffield S10 2TN, United Kingdom. P.Baker@sheffield.ac.uk