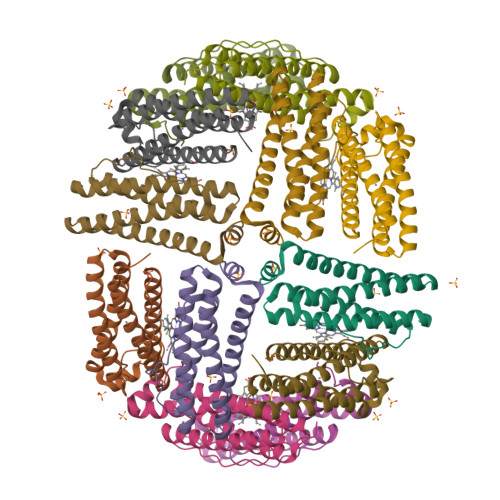
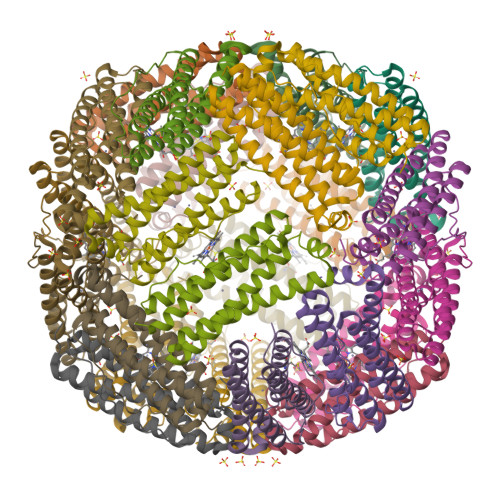
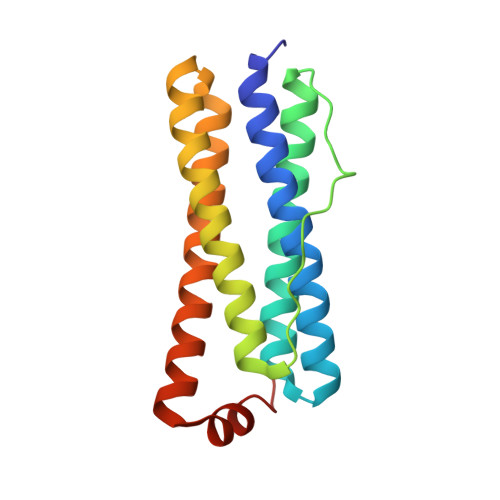
The Binding of Haem and Zinc in the 1.9 A X-Ray Structure of Escherichia Coli Bacterioferritin.
Willies, S.C., Isupov, M.N., Garman, E.F., Littlechild, J.A.(2009) J Biol Inorg Chem 14: 201
- PubMed: 18946693
- DOI: https://doi.org/10.1007/s00775-008-0438-8
- Primary Citation of Related Structures:
2VXI - PubMed Abstract:
The crystal structure of Escherichia coli bacterioferritin has been solved to 1.9 A, and shows the symmetrical binding of a haem molecule on the local twofold axis between subunits and a pair of metal atoms bound to each subunit at the ferroxidase centre. These metals have been identified as zinc by the analysis of the structure and X-ray data and confirmed by microfocused proton-induced X-ray emission experiments. For the first time the haem has been shown to be linked to both the internal and the external environments via a cluster of waters positioned above the haem molecule.
Organizational Affiliation:
School of Biosciences, Henry Wellcome Building for Biocatalysis, University of Exeter, Stocker Road, Exeter, EX4 4QD, UK.