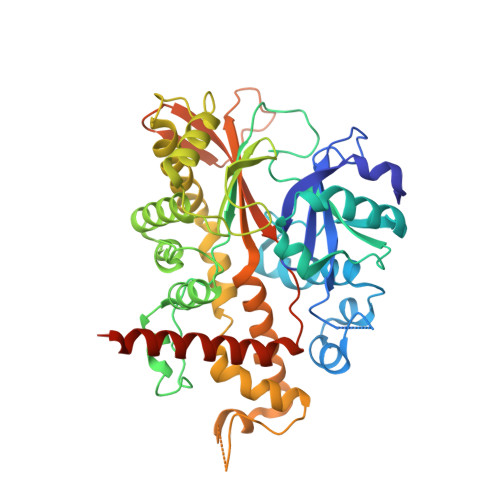
Inositol 1,3,4,5,6-Pentakisphosphate 2-Kinase is a Distant Ipk Member with a Singular Inositide Binding Site for Axial 2-Oh Recognition.
Gonzalez, B., Banos-Sanz, J.I., Villate, M., Brearley, C.A., Sanz-Aparicio, J.(2010) Proc Natl Acad Sci U S A 107: 9608
- PubMed: 20453199
- DOI: https://doi.org/10.1073/pnas.0912979107
- Primary Citation of Related Structures:
2XAL, 2XAM, 2XAN, 2XAO, 2XAR - PubMed Abstract:
Inositol phosphates (InsPs) are signaling molecules with multiple roles in cells. In particular (InsP(6)) is involved in mRNA export and editing or chromatin remodeling among other events. InsP(6) accumulates as mixed salts (phytate) in storage tissues of plants and plays a key role in their physiology. Human diets that are exclusively grain-based provide an excess of InsP(6) that, through chelation of metal ions, may have a detrimental effect on human health. Ins(1,3,4,5,6)P(5) 2-kinase (InsP(5) 2-kinase or Ipk1) catalyses the synthesis of InsP(6) from InsP(5) and ATP, and is the only enzyme that transfers a phosphate group to the axial 2-OH of the myo-inositide. We present the first structure for an InsP(5) 2-kinase in complex with both substrates and products. This enzyme presents a singular structural region for inositide binding that encompasses almost half of the protein. The key residues in substrate binding are identified, with Asp368 being responsible for recognition of the axial 2-OH. This study sheds light on the unique molecular mechanism for the synthesis of the precursor of inositol pyrophosphates.
Organizational Affiliation:
Grupo de Cristalografía Macromolecular y Biología Estructural, Instituto de Química-Física Rocasolano, Consejo Superior de Investigaciones Científicas, Serrano 119, 28006 Madrid, Spain. xbeatriz@iqfr.csic.es