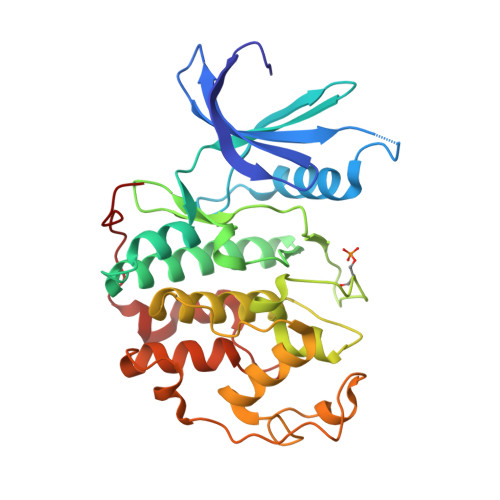
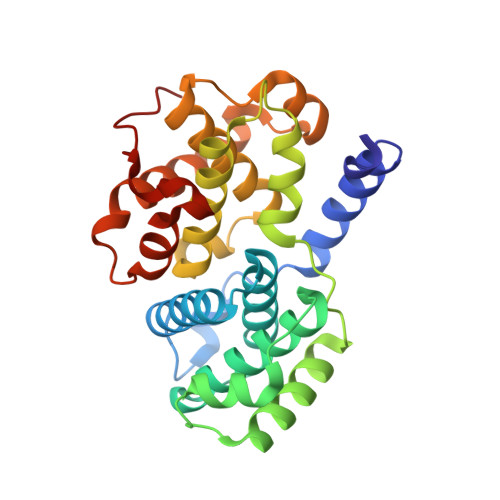
Meriolins, a new class of cell death inducing kinase inhibitors with enhanced selectivity for cyclin-dependent kinases
Bettayeb, K., Tirado, O.M., Marionneau-Lambot, S., Ferandin, Y., Lozach, O., Morris, J.C., Mateo-Lozano, S., Drueckes, P., Kubbutat, M.H., Liger, F., Marquet, B., Joseph, B., Echalier, A., Endicott, J.A., Notario, V., Meijer, L.(2007) Cancer Res 67: 8325-8334
- PubMed: 17804748
- DOI: https://doi.org/10.1158/0008-5472.CAN-07-1826
- Primary Citation of Related Structures:
3BHT, 3BHU, 3BHV - PubMed Abstract:
Protein kinases represent promising anticancer drug targets. We describe here the meriolins, a new family of inhibitors of cyclin-dependent kinases (CDK). Meriolins represent a chemical structural hybrid between meridianins and variolins, two families of kinase inhibitors extracted from various marine invertebrates. Variolin B is currently in preclinical evaluation as an antitumor agent. A selectivity study done on 32 kinases showed that, compared with variolin B, meriolins display enhanced specificity toward CDKs, with marked potency on CDK2 and CDK9. The structures of pCDK2/cyclin A/variolin B and pCDK2/cyclin A/meriolin 3 complexes reveal that the two inhibitors bind within the ATP binding site of the kinase, but in different orientations. Meriolins display better antiproliferative and proapoptotic properties in human tumor cell cultures than their parent molecules, meridianins and variolins. Phosphorylation at CDK1, CDK4, and CDK9 sites on, respectively, protein phosphatase 1alpha, retinoblastoma protein, and RNA polymerase II is inhibited in neuroblastoma SH-SY5Y cells exposed to meriolins. Apoptosis triggered by meriolins is accompanied by rapid Mcl-1 down-regulation, cytochrome c release, and activation of caspases. Meriolin 3 potently inhibits tumor growth in two mouse xenograft cancer models, namely, Ewing's sarcoma and LS174T colorectal carcinoma. Meriolins thus constitute a new CDK inhibitory scaffold, with promising antitumor activity, derived from molecules initially isolated from marine organisms.
Organizational Affiliation:
Centre National de la Recherche Scientifique, Cell Cycle Group & UPS2682, Station Biologique, Roscoff, Bretagne, France.