Insights into base selectivity from the structures of an RB69 DNA Polymerase triple mutant
Klimenko, D., Wang, M., Steitz, T.A., Konigsberg, W.H., Wang, J.To be published.
Experimental Data Snapshot
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| DNA polymerase | C [auth A] | 909 | N/A | Mutation(s): 3 Gene Names: 43 EC: 2.7.7.7 (PDB Primary Data), 3.1.11 (UniProt) | 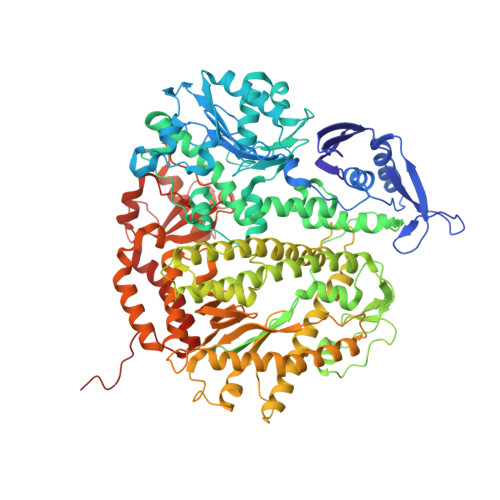 |
UniProt | |||||
Find proteins for Q38087 (Escherichia phage RB69) Explore Q38087 Go to UniProtKB: Q38087 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q38087 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*DAP*DCP*DAP*DGP*DGP*DTP*DAP*DAP*DGP*DCP*DAP*DGP*DTP*DCP*DCP*DGP*DCP*DG)-3') | A [auth T] | 18 | N/A | 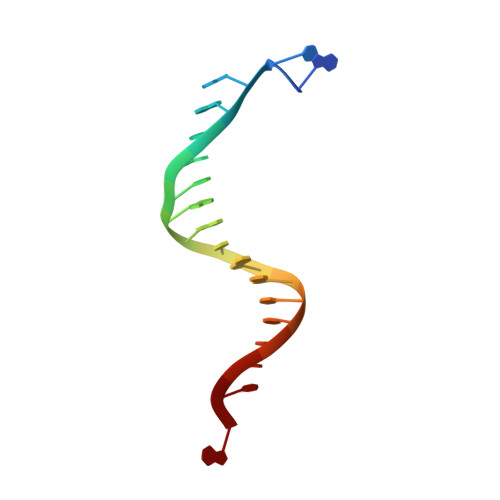 | |
Sequence AnnotationsExpand | |||||
| |||||
Find similar nucleic acids by: Sequence | 3D Structure
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Length | Organism | Image | |
| DNA (5'-D(*DGP*DCP*DGP*DGP*DAP*DCP*DTP*DGP*DCP*DTP*DTP*DAP*DCP*(DOC))-3') | B [auth P] | 14 | N/A | 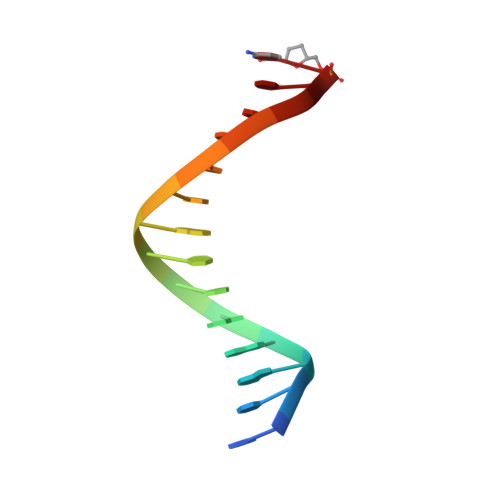 | |
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 3 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| TTP Query on TTP | I [auth A] | THYMIDINE-5'-TRIPHOSPHATE C10 H17 N2 O14 P3 NHVNXKFIZYSCEB-XLPZGREQSA-N |  | ||
| CA Query on CA | D [auth A], E [auth A], F [auth A], G [auth A] | CALCIUM ION Ca BHPQYMZQTOCNFJ-UHFFFAOYSA-N |  | ||
| CL Query on CL | H [auth A] | CHLORIDE ION Cl VEXZGXHMUGYJMC-UHFFFAOYSA-M |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 80.726 | α = 90 |
| b = 117.281 | β = 90 |
| c = 126.813 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data collection |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| CNS | phasing |