Flexible cyclic ethers/polyethers as novel P2-ligands for HIV-1 protease inhibitors: design, synthesis, biological evaluation, and protein-ligand X-ray studies
Ghosh, A.K., Gemma, S., Baldridge, A., Wang, Y.F., Kovalevsky, A.Y., Koh, Y., Weber, I.T., Mitsuya, H.(2008) J Med Chem 51: 6021-6033
- PubMed: 18783203
- DOI: https://doi.org/10.1021/jm8004543
- Primary Citation of Related Structures:
3DJK - PubMed Abstract:
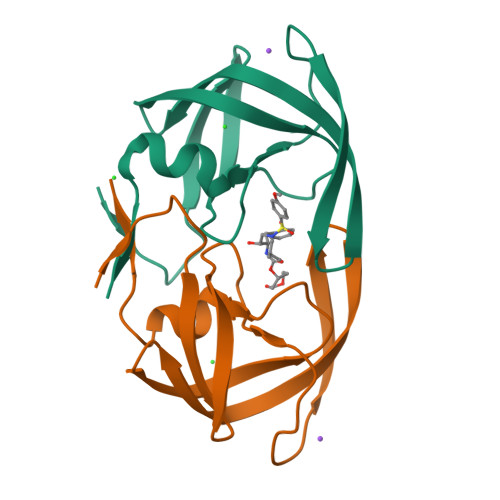
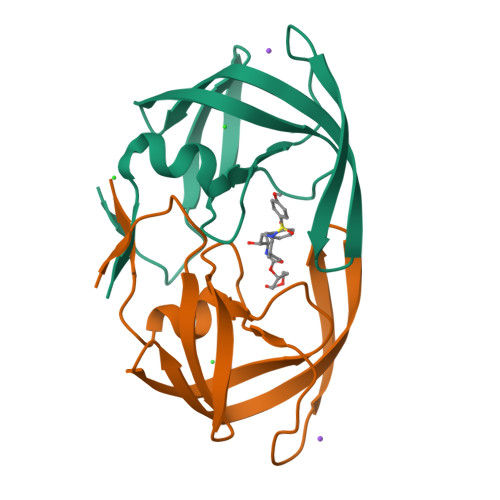
We report the design, synthesis, and biological evaluation of a series of novel HIV-1 protease inhibitors. The inhibitors incorporate stereochemically defined flexible cyclic ethers/polyethers as high affinity P2-ligands. Inhibitors containing small ring 1,3-dioxacycloalkanes have shown potent enzyme inhibitory and antiviral activity. Inhibitors 3d and 3h are the most active inhibitors. Inhibitor 3d maintains excellent potency against a variety of multi-PI-resistant clinical strains. Our structure-activity studies indicate that the ring size, stereochemistry, and position of oxygens are important for the observed activity. Optically active synthesis of 1,3-dioxepan-5-ol along with the syntheses of various cyclic ether and polyether ligands have been described. A protein-ligand X-ray crystal structure of 3d-bound HIV-1 protease was determined. The structure revealed that the P2-ligand makes extensive interactions including hydrogen bonding with the protease backbone in the S2-site. In addition, the P2-ligand in 3d forms a unique water-mediated interaction with the NH of Gly-48.
Organizational Affiliation:
Department of Chemistry, Purdue University, West Lafayette, Indiana 47907, USA. akghosh@purdue.edu