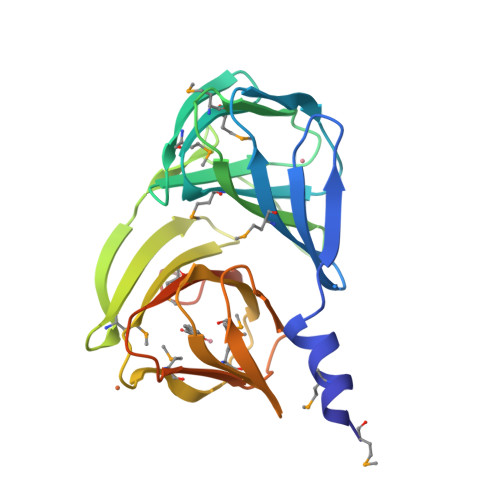
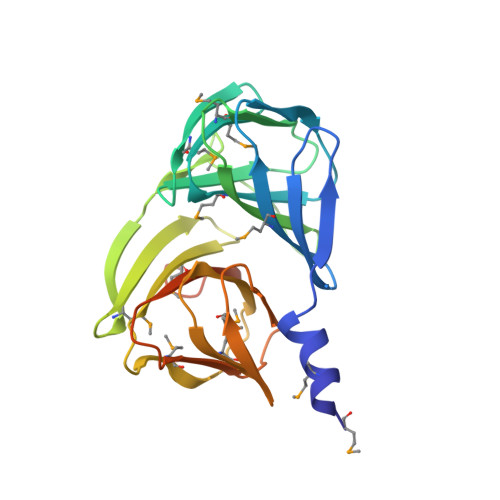
Analysis of multiple crystal forms of Bacillus subtilis BacB suggests a role for a metal ion as a nucleant for crystallization
Rajavel, M., Gopal, B.(2010) Acta Crystallogr D Biol Crystallogr 66: 635-639
- PubMed: 20445239
- DOI: https://doi.org/10.1107/S0907444910006682
- Primary Citation of Related Structures:
3H9A - PubMed Abstract:
Bacillus subtilis BacB is an oxidase that is involved in the production of the antibiotic bacilysin. This protein contains two double-stranded beta-helix (cupin) domains fused in a compact arrangement. BacB crystallizes in three crystal forms under similar crystallization conditions. An interesting observation was that a slight perturbation of the crystallization droplet resulted in the nucleation of a different crystal form. An X-ray absorption scan of BacB suggested the presence of cobalt and iron in the crystal. Here, a comparative analysis of the different crystal forms of BacB is presented in an effort to identify the basis for the different lattices. It is noted that metal ions mediating interactions across the asymmetric unit dominate the different packing arrangements. Furthermore, a normalized B-factor analysis of all the crystal structures suggests that the solvent-exposed metal ions decrease the flexibility of a loop segment, perhaps influencing the choice of crystal form. The residues coordinating the surface metal ion are similar in the triclinic and monoclinic crystal forms. The coordinating ligands for the corresponding metal ion in the tetragonal crystal form are different, leading to a tighter packing arrangement. Although BacB is a monomer in solution, a dimer of BacB serves as a template on which higher order symmetrical arrangements are formed. The different crystal forms of BacB thus provide experimental evidence for metal-ion-mediated lattice formation and crystal packing.
Organizational Affiliation:
Molecular Biophysics Unit, Indian Institute of Science, Bangalore 560 012, India.