Biocatalysis with thermostable enzymes: structure and properties of a thermophilic 'ene'-reductase related to old yellow enzyme.
Adalbjornsson, B.V., Toogood, H.S., Fryszkowska, A., Pudney, C.R., Jowitt, T.A., Leys, D., Scrutton, N.S.(2010) Chembiochem 11: 197-207
- PubMed: 19943268
- DOI: https://doi.org/10.1002/cbic.200900570
- Primary Citation of Related Structures:
3KRU, 3KRZ - PubMed Abstract:
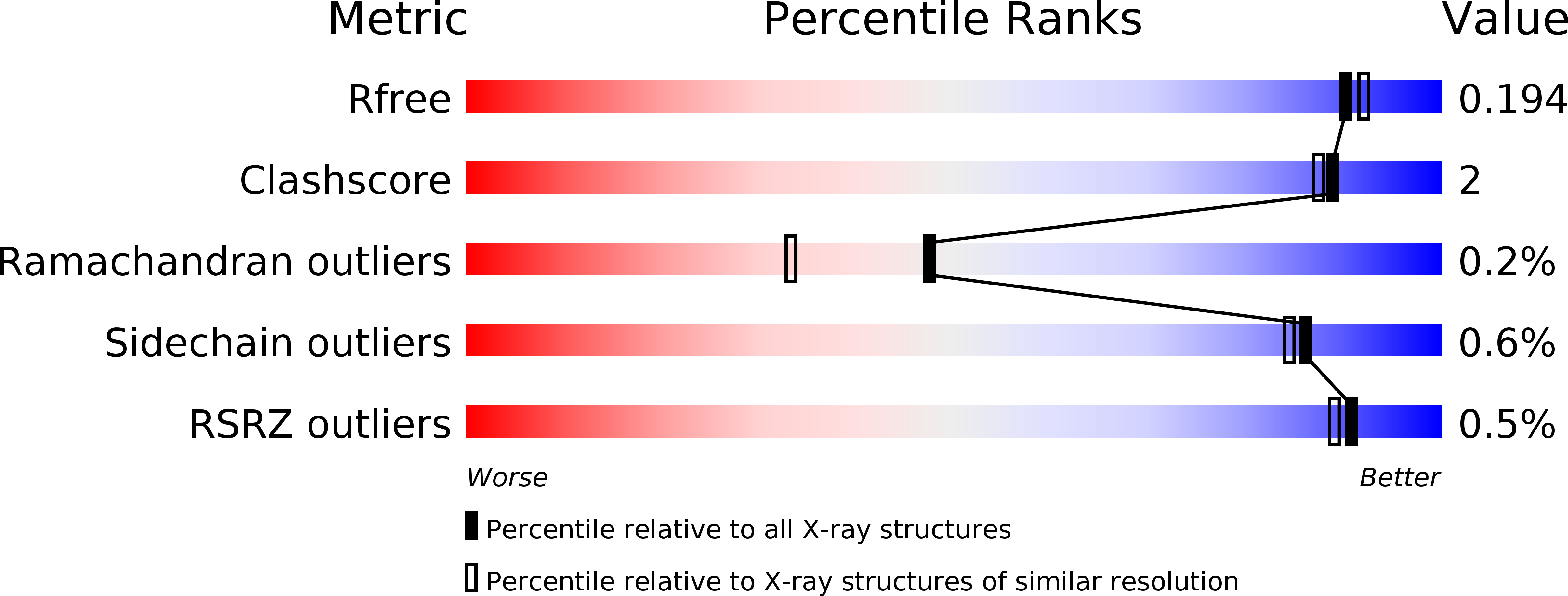
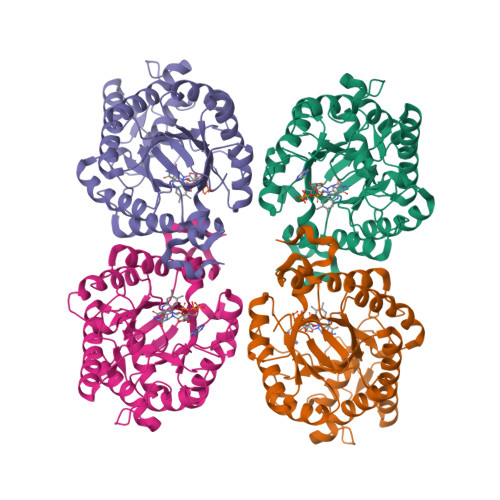
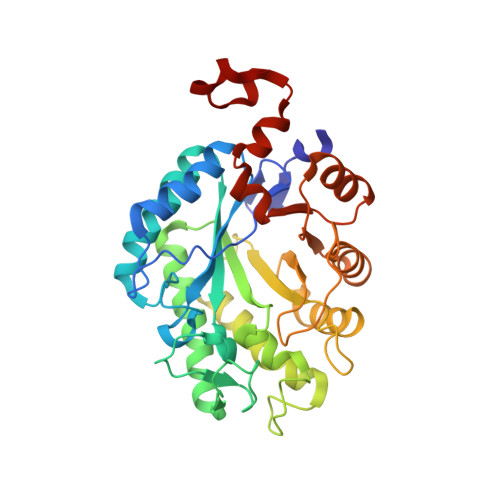
We report the crystal structure of a thermophilic "ene" reductase (TOYE) isolated from Thermoanaerobacter pseudethanolicus E39. The crystal structure reveals a tetrameric enzyme and an active site that is relatively large compared to most other structurally determined and related Old Yellow Enzymes. The enzyme adopts higher order oligomeric states (octamers and dodecamers) in solution, as revealed by sedimentation velocity and multiangle laser light scattering. Bead modelling indicates that the solution structure is consistent with the basic tetrameric structure observed in crystallographic studies and electron microscopy. TOYE is stable at high temperatures (T(m)>70 degrees C) and shows increased resistance to denaturation in water-miscible organic solvents compared to the mesophilic Old Yellow Enzyme family member, pentaerythritol tetranitrate reductase. TOYE has typical ene-reductase properties of the Old Yellow Enzyme family. There is currently major interest in using Old Yellow Enzyme family members in the preparative biocatalysis of a number of activated alkenes. The increased stability of TOYE in organic solvents is advantageous for biotransformations in which water-miscible organic solvents and biphasic reaction conditions are required to both deliver novel substrates and minimize product racemisation.
Organizational Affiliation:
Faculty of Life Sciences, University of Manchester, 131 Princess Street, Manchester M1 7DN, UK.