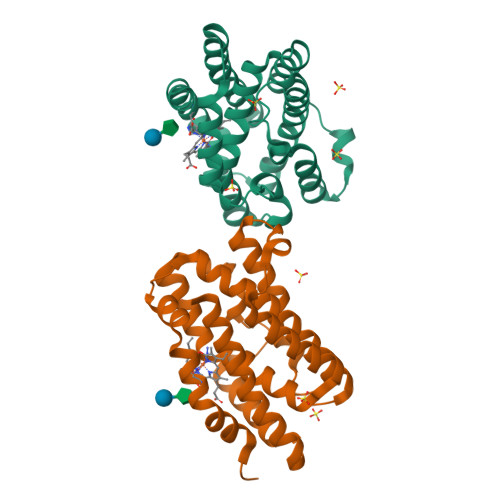
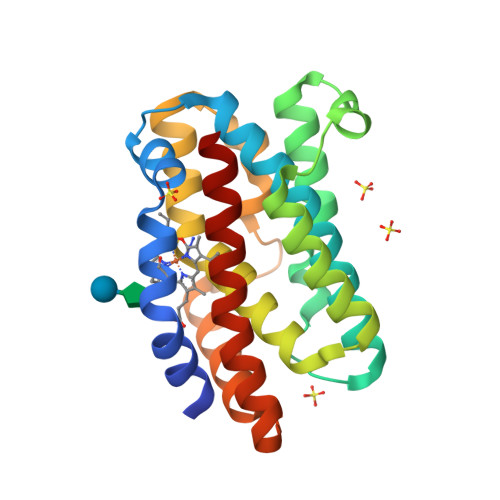
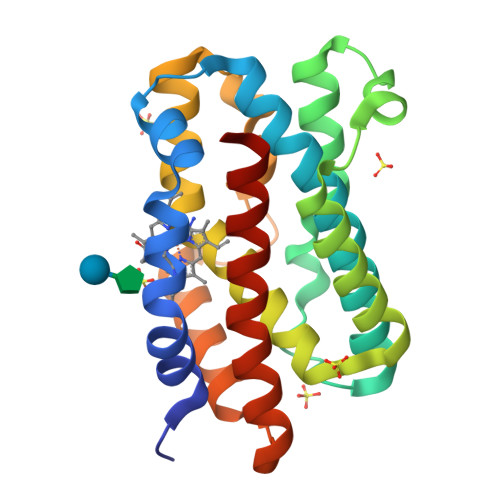
Enzymatic ring-opening mechanism of verdoheme by the heme oxygenase: a combined X-ray crystallography and QM/MM study.
Lai, W., Chen, H., Matsui, T., Omori, K., Unno, M., Ikeda-Saito, M., Shaik, S.(2010) J Am Chem Soc 132: 12960-12970
- PubMed: 20806922
- DOI: https://doi.org/10.1021/ja104674q
- Primary Citation of Related Structures:
3MOO - PubMed Abstract:
The least understood mechanism during heme degradation by the enzyme heme oxygenase (HO) is the third step of ring opening of verdoheme to biliverdin, a process which maintains iron homeostasis. In response to this mechanistic uncertainty, we launched a combined study of X-ray crystallography and theoretical QM/MM calculations, designed to elucidate the mechanism. The air-sensitive ferrous verdoheme complex of HmuO, a heme oxygenase from Corynebacterium diphtheriae, was crystallized under anaerobic conditions. Spectral analysis of the azide-bound verdoheme-HmuO complex crystals assures that the verdoheme group remains intact during the crystallization and X-ray diffraction measurement. The structure offers the first solid evidence for the presence of a water cluster in the distal pocket of this catalytically critical intermediate. The subsequent QM/MM calculations based on this crystal structure explore the reaction mechanisms starting from the FeOOH-verdoheme and FeHOOH-verdoheme complexes, which mimic, respectively, the O(2)- and H(2)O(2)-supported degradations. In both mechanisms, the rate-determining step is the initial O-O bond breaking step, which is either homolytic (for FeHOOH-verdoheme) or coupled to electron and proton transfers (in FeOOH-verdoheme). Additionally, the calculations indicate that the FeHOOH-verdoheme complex is more reactive than the FeOOH-verdoheme complex in accord with experimental findings. QM energies with embedded MM charges are close to and yield the same conclusions as full QM/MM energies. Finally, the calculations highlight the dominant influence of the distal water cluster which acts as a biocatalyst for the conversion of verdoheme to biliverdin in the two processes, by fixing the departing OH and directing it to the requisite site of attack, and by acting as a proton shuttle and a haven for the highly reactive OH(-) nucleophile.
- Institute of Chemistry and The Lise Meitner-Minerva Center for Computational Quantum Chemistry, The Hebrew University of Jerusalem, 91904 Jerusalem, Israel.
Organizational Affiliation: