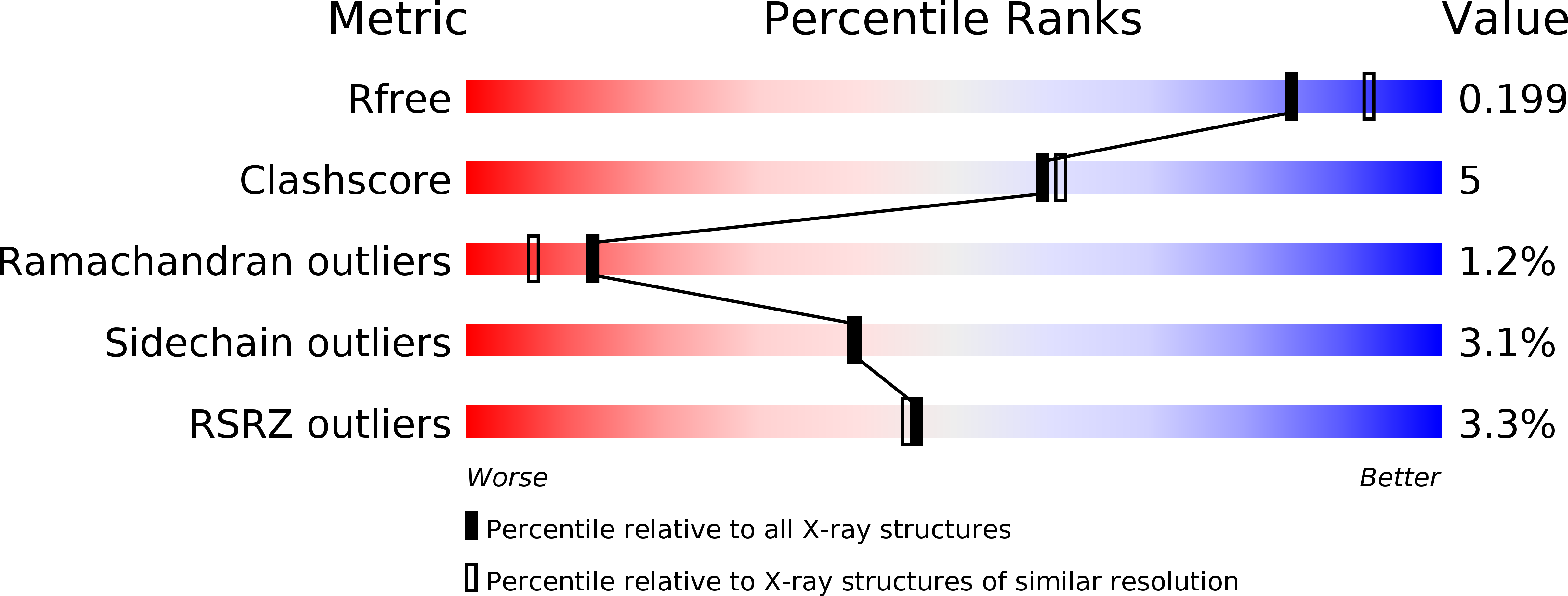
Probing the Conformational Mobility of the Active Site of Ascorbate Peroxidase
Guimero, A., Badyal, S.K., Leeks, T., Moody, P.C.E., Raven, E.L.(2013) Dalton Trans 42: 3170
- PubMed: 23202589
- DOI: https://doi.org/10.1039/c2dt32455e
- Primary Citation of Related Structures:
3ZCG, 3ZCH, 3ZCY - PubMed Abstract:
We have previously demonstrated (Badyal et al., J. Biol. Chem., 2006, 281, 24512) that removal of the active site tryptophan (Trp41) in ascorbate peroxidase increases the conformational mobility of the distal histidine residue (His42) and that His42 coordinates to the iron in the oxidised W41A enzyme to give a 6-coordinate, low-spin peroxidase. In this work, we probe the conformational flexibility of the active site in more detail. We examine whether other residues (Cys, Tyr, Met) can also ligate to the heme at position 42; we find that introduction of other ligating amino acids created a cavity in the heme pocket, but that formation of 6-coordinate heme is not observed. In addition, we examine the role of Asn-71, which hydrogen bonds to His42 and tethers the distal histidine in the active site pocket; we find that removal of this hydrogen bond increases the proportion of low-spin heme. We suggest that, in addition to its well-known role in facilitating the reaction with peroxide, His42 also plays a role in defining the shape and folding of the active site pocket.