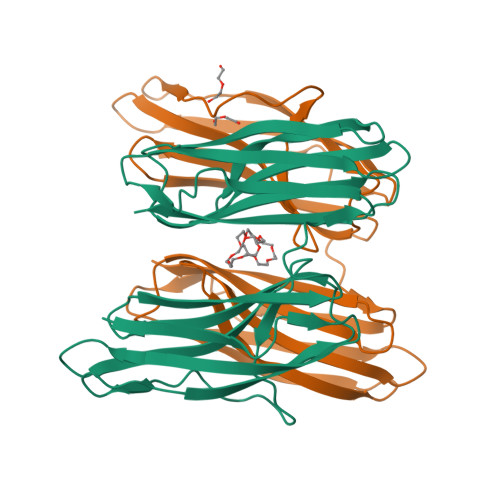
Structural Insights Into Rbma, a Biofilm Scaffolding Protein of V. Cholerae.
Maestre-Reyna, M., Wu, W., Wang, A.H.(2013) PLoS One 8: 82458
- PubMed: 24340031
- DOI: https://doi.org/10.1371/journal.pone.0082458
- Primary Citation of Related Structures:
4BE5, 4BE6, 4BEI - PubMed Abstract:
V. cholerae can form sessile biofilms associated with abiotic surfaces, cyanobacteria, zoo-plankton, mollusks, or crustaceans. Along with the vibrio polysaccharide, secreted proteins of the rbm gene cluster are key to the biofilm ultrastructure. Here we provide a thorough structural characterization of RbmA, a protein involved in mediating cell-cell and cell-biofilm contacts. We correlate our structural findings with initial ligand specificity screening results, NMR protein-ligand interaction analysis, and complement our results with a full biocomputational study.
Organizational Affiliation:
Institute of Biological Chemistry, Academia Sinica, Taipei, Taiwan.