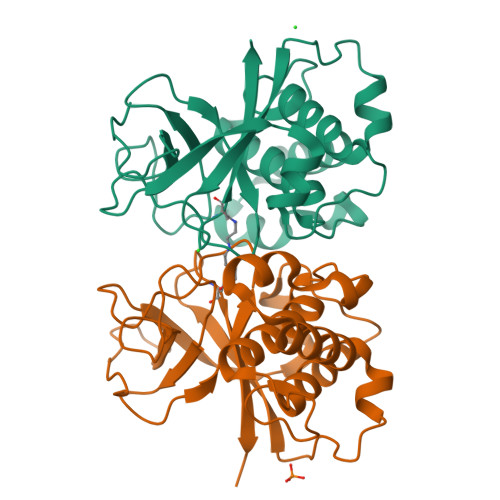
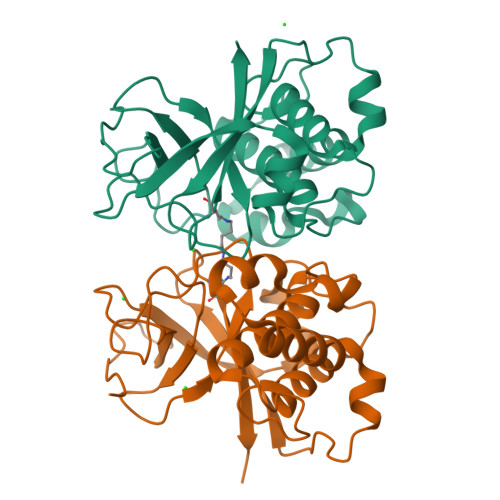
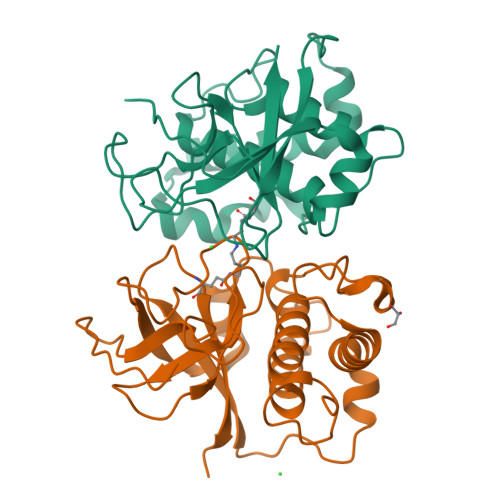
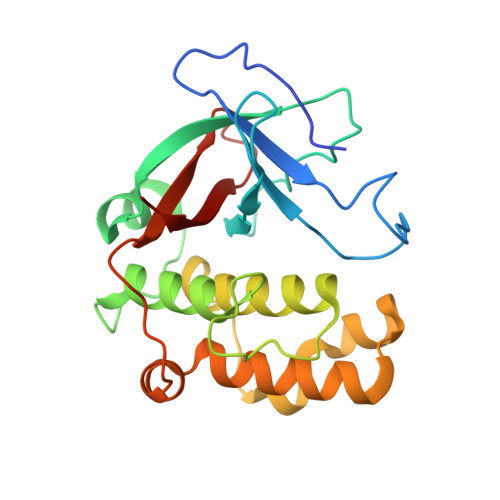
Structural Model for the Covalent Adhesion of the Streptococcus Pyogenes Pilus Through a Thioester Bond.
Linke-Winnebeck, C., Paterson, N.G., Young, P.G., Middleditch, M.J., Greenwood, D.R., Witte, G., Baker, E.N.(2014) J Biological Chem 289: 177
- PubMed: 24220033
- DOI: https://doi.org/10.1074/jbc.M113.523761
- Primary Citation of Related Structures:
4C0Z - PubMed Abstract:
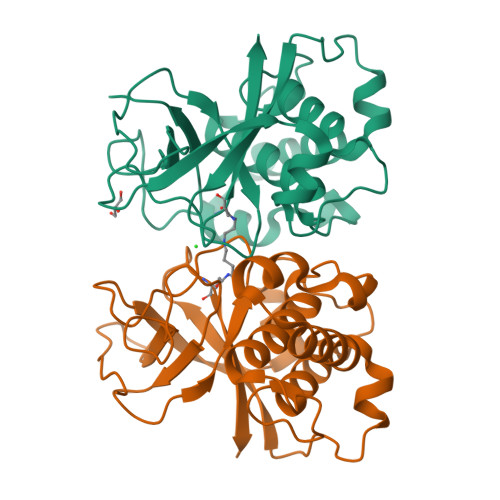
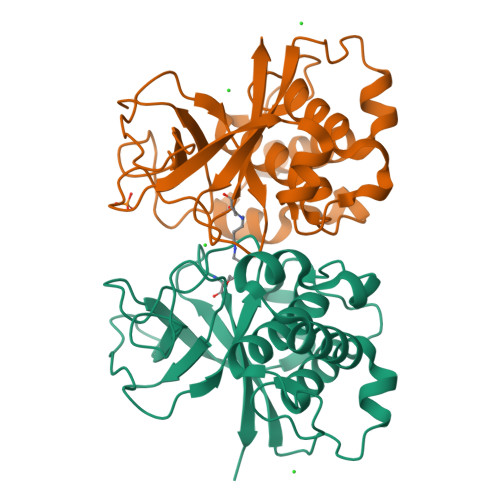
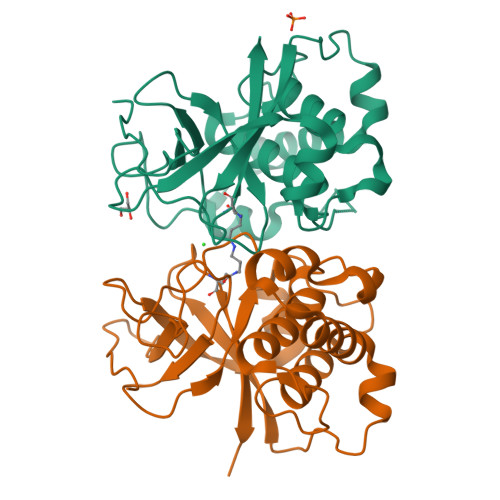
The human pathogen Streptococcus pyogenes produces pili that are essential for adhesion to host surface receptors. Cpa, the adhesin at the pilus tip, was recently shown to have a thioester-containing domain. The thioester bond is believed to be important in adhesion, implying a mechanism of covalent attachment analogous to that used by human complement factors. Here, we have characterized a second active thioester-containing domain on Cpa, the N-terminal domain of Cpa (CpaN). Expression of CpaN in Escherichia coli gave covalently linked dimers. These were shown by x-ray crystallography and mass spectrometry to comprise two CpaN molecules cross-linked by the polyamine spermidine following reaction with the thioester bonds. This cross-linked CpaN dimer provides a model for the covalent attachment of Cpa to target receptors and thus the streptococcal pilus to host cells. Similar thioester domains were identified in cell wall proteins of other Gram-positive pathogens, suggesting that thioester domains are more widely used and provide a mechanism of adhesion by covalent bonding to target molecules on host cells that mimics that used by the human complement system to eliminate pathogens.
Organizational Affiliation:
From the School of Biological Sciences and Maurice Wilkins Centre for Molecular Biodiscovery, University of Auckland, Private Bag 921019, Auckland 1142, New Zealand and.