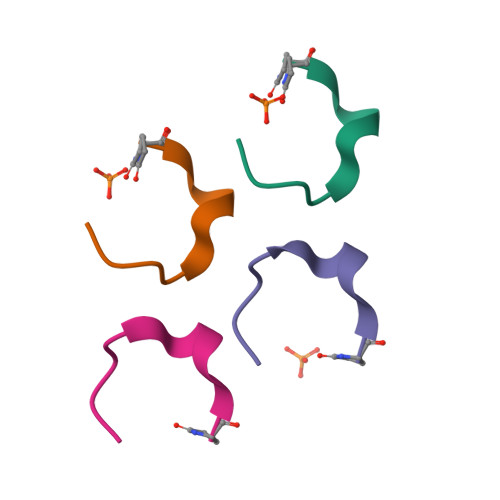
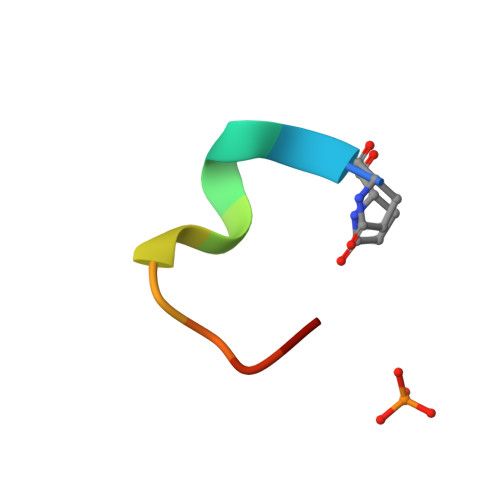
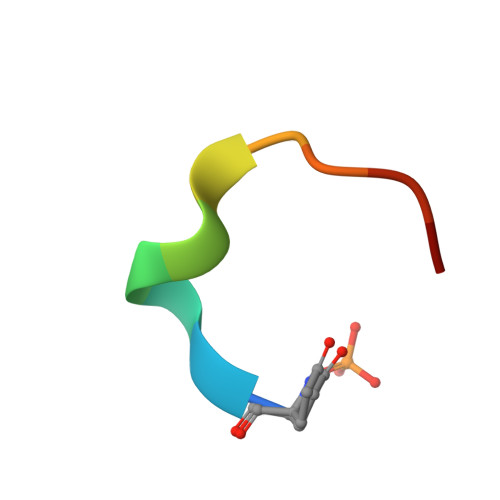
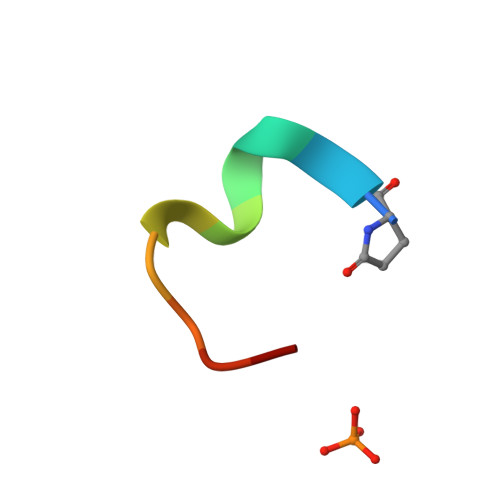
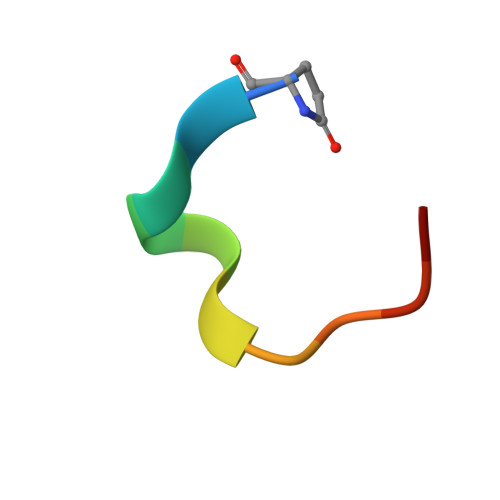
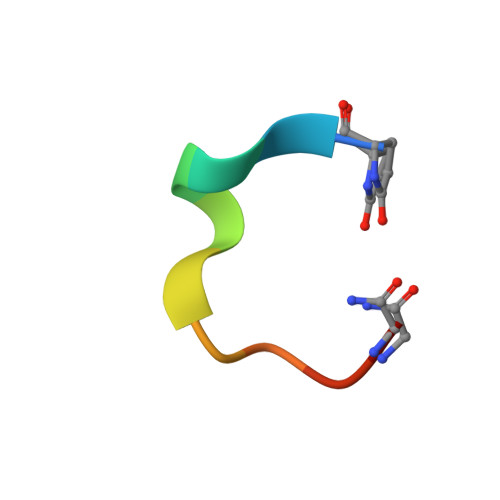
Atomic View of the Histidine Environment Stabilizing Higher- Ph Conformations of Ph-Dependent Proteins.
Valery, C., Deville-Foillard, S., Lefebvre, C., Taberner, N., Legrand, P., Meneau, F., Meriadec, C., Delvaux, C., Bizien, T., Kasotakis, E., Lopez-Iglesias, C., Gall, A., Bressanelli, S., Le Du, M.-H., Paternostre, M., Artzner, F.(2015) Nat Commun 6: 7771
- PubMed: 26190377
- DOI: https://doi.org/10.1038/ncomms8771
- Primary Citation of Related Structures:
4D5M - PubMed Abstract:
External stimuli are powerful tools that naturally control protein assemblies and functions. For example, during viral entry and exit changes in pH are known to trigger large protein conformational changes. However, the molecular features stabilizing the higher pH structures remain unclear. Here we elucidate the conformational change of a self-assembling peptide that forms either small or large nanotubes dependent on the pH. The sub-angstrom high-pH peptide structure reveals a globular conformation stabilized through a strong histidine-serine H-bond and a tight histidine-aromatic packing. Lowering the pH induces histidine protonation, disrupts these interactions and triggers a large change to an extended β-sheet-based conformation. Re-visiting available structures of proteins with pH-dependent conformations reveals both histidine-containing aromatic pockets and histidine-serine proximity as key motifs in higher pH structures. The mechanism discovered in this study may thus be generally used by pH-dependent proteins and opens new prospects in the field of nanomaterials.
Organizational Affiliation:
1] Biomolecular Interaction Centre, School of Biological Sciences, University of Canterbury, 8140 Christchurch, New zealand [2] Ipsen, 5 Avenue du Canada, 91940 Les Ulis, France.