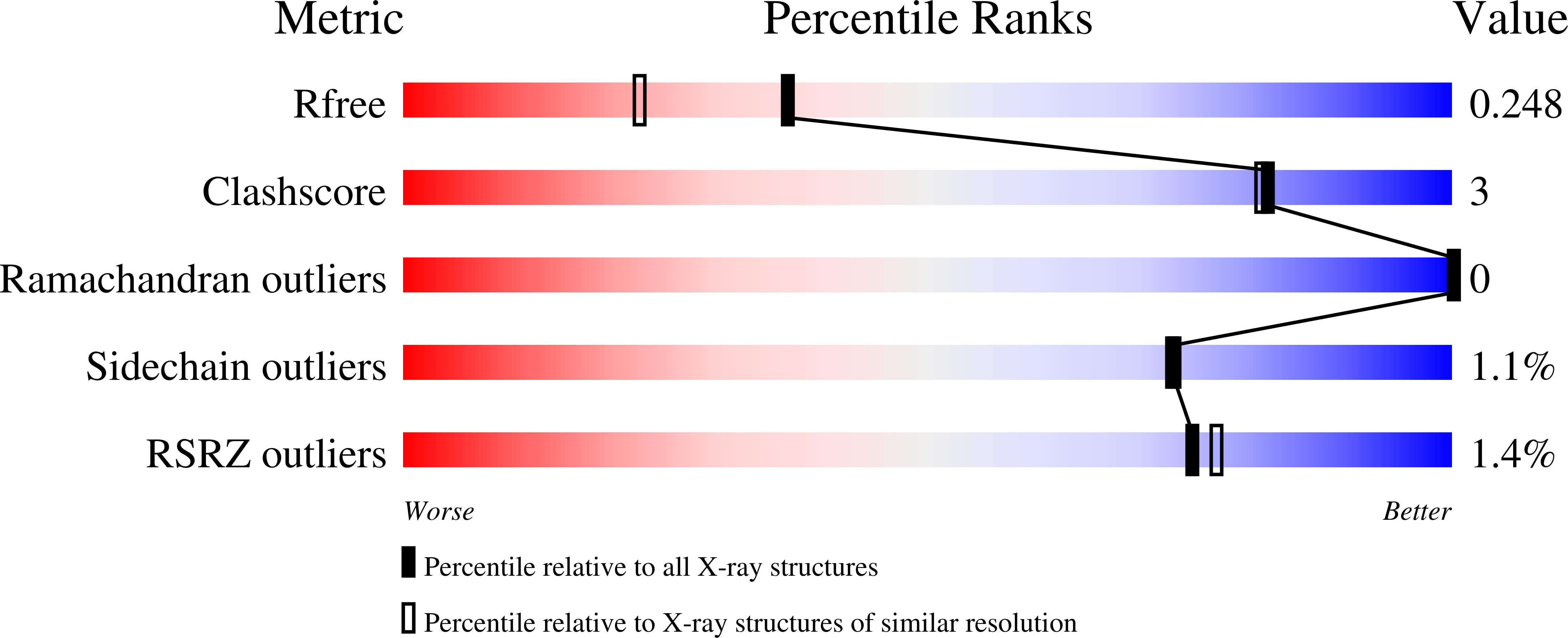
Structural Insights Into the Inhibition of Cellobiohydrolase Cel7A by Xylooligosaccharides.
Haddad Momeni, M., Ubhayasekera, W., Sandgren, M., Stahlberg, J., Hansson, H.(2015) FEBS J 282: 2167
- PubMed: 25765184
- DOI: https://doi.org/10.1111/febs.13265
- Primary Citation of Related Structures:
4D5I, 4D5J, 4D5O, 4D5P, 4D5Q, 4D5V - PubMed Abstract:
The filamentous fungus Hypocrea jecorina (anamorph of Trichoderma reesei) is the predominant source of enzymes for industrial saccharification of lignocellulose biomass. The major enzyme, cellobiohydrolase Cel7A, constitutes nearly half of the total protein in the secretome. The performance of such enzymes is susceptible to inhibition by compounds liberated by physico-chemical pre-treatment if the biomass is kept unwashed. Xylan and xylo-oligosaccharides (XOS) have been proposed to play a key role in inhibition of cellobiohydrolases of glycoside hydrolase family 7. To elucidate the mechanism behind this inhibition at a molecular level, we used X-ray crystallography to determine structures of H. jecorina Cel7A in complex with XOS. Structures with xylotriose, xylotetraose and xylopentaose revealed a predominant binding mode at the entrance of the substrate-binding tunnel of the enzyme, in which each xylose residue is shifted ~ 2.4 Å towards the catalytic center compared with binding of cello-oligosaccharides. Furthermore, partial occupancy of two consecutive xylose residues at subsites -2 and -1 suggests an alternative binding mode for XOS in the vicinity of the catalytic center. Interestingly, the -1 xylosyl unit exhibits an open aldehyde conformation in one of the structures and a ring-closed pyranoside in another complex. Complementary inhibition studies with p-nitrophenyl lactoside as substrate indicate mixed inhibition rather than pure competitive inhibition. The atomic coordinates and structure factors are available in the Protein Data Bank under accession number 4D5I (H. jecorina Cel7A E212Q variant, complex with xylotriose), 4D5J (H. jecorina Cel7A E217Q variant, complex with xylotriose), 4D5O (H. jecorina Cel7A E212Q variant, complex with xylopentaose), 4D5P (H. jecorina Cel7A E217Q variant, complex with xylopentaose), 4D5Q (wild-type H. jecorina Cel7A, complex with xylopentaose) and 4D5V (H. jecorina Cel7A E217Q variant, complex with xylotetraose).
Organizational Affiliation:
Department of Chemistry and Biotechnology, Swedish University of Agricultural Sciences, Uppsala, Sweden.