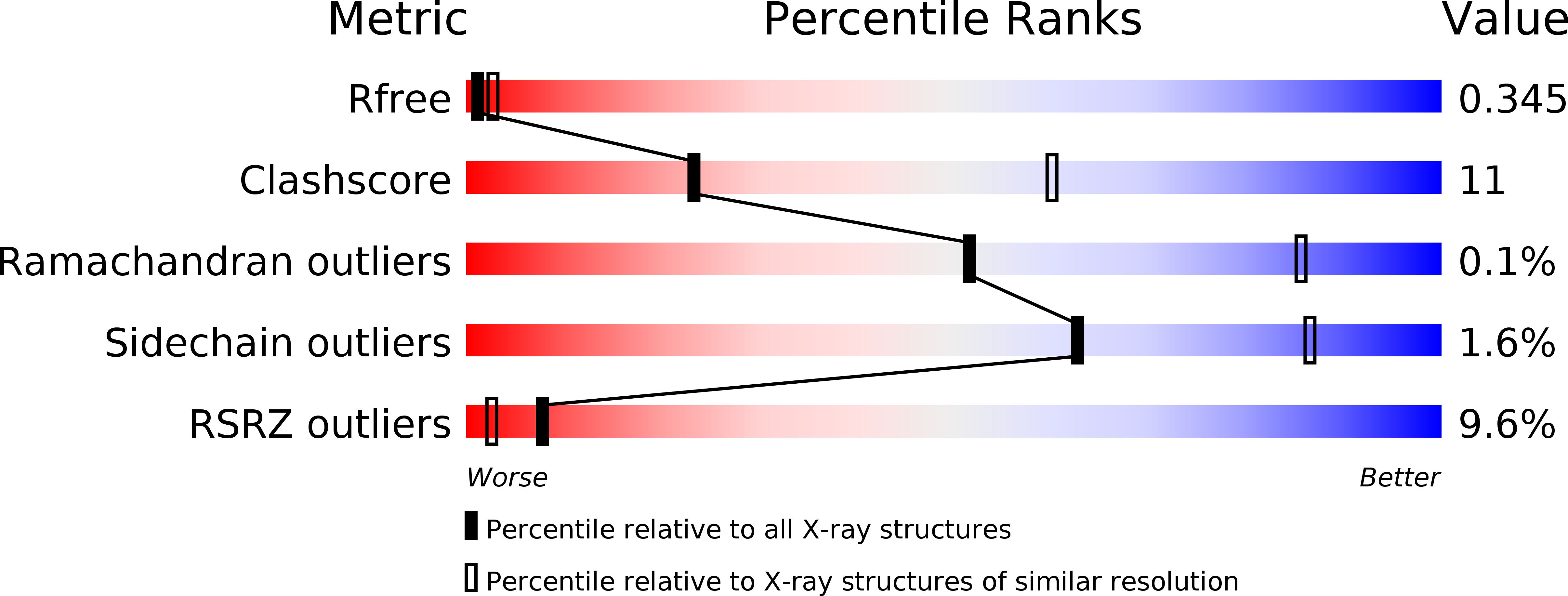
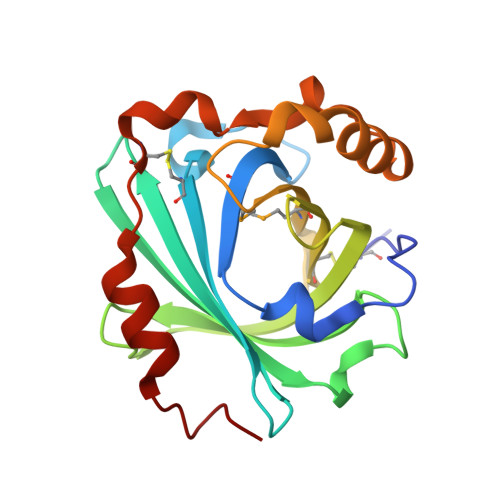
Structure and ligand-binding properties of the biogenic amine-binding protein from the saliva of a blood-feeding insect vector of Trypanosoma cruzi.
Xu, X., Chang, B.W., Mans, B.J., Ribeiro, J.M., Andersen, J.F.(2013) Acta Crystallogr D Biol Crystallogr 69: 105-113
- PubMed: 23275168
- DOI: https://doi.org/10.1107/S0907444912043326
- Primary Citation of Related Structures:
4GE1, 4GET, 4HFO - PubMed Abstract:
Proteins that bind small-molecule mediators of inflammation and hemostasis are essential for blood-feeding by arthropod vectors of infectious disease. In ticks and triatomine insects, the lipocalin protein family is greatly expanded and members have been shown to bind biogenic amines, eicosanoids and ADP. These compounds are potent mediators of platelet activation, inflammation and vascular tone. In this paper, the structure of the amine-binding protein (ABP) from Rhodnius prolixus, a vector of the trypanosome that causes Chagas disease, is described. ABP binds the biogenic amines serotonin and norepinephrine with high affinity. A complex with tryptamine shows the presence of a binding site for a single ligand molecule in the central cavity of the β-barrel structure. The cavity contains significant additional volume, suggesting that this protein may have evolved from the related nitrophorin proteins, which bind a much larger heme ligand in the central cavity.
Organizational Affiliation:
Laboratory of Malaria and Vector Research, NIH/NIAID, Rockville, MD 20852, USA.