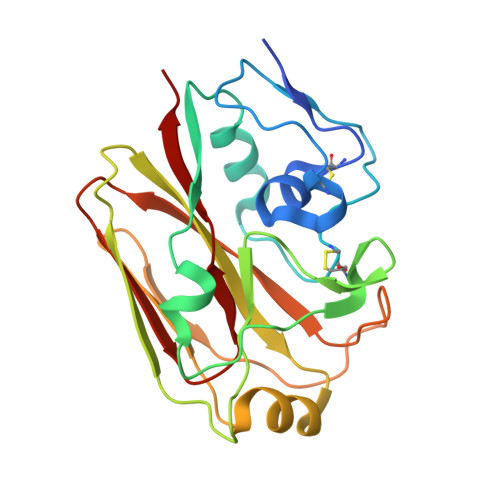
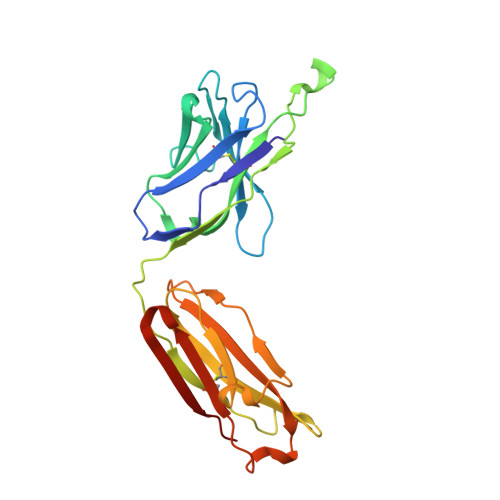
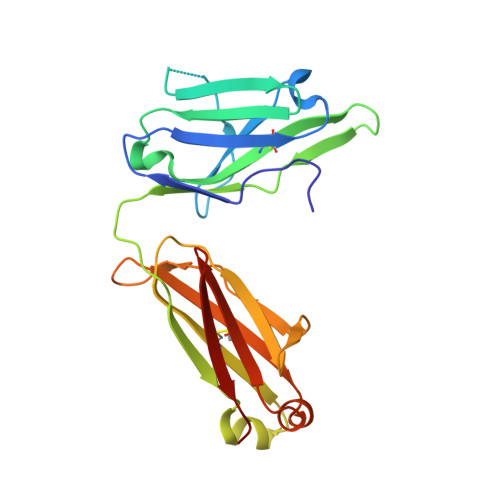
Preconfiguration of the antigen-binding site during affinity maturation of a broadly neutralizing influenza virus antibody.
Schmidt, A.G., Xu, H., Khan, A.R., O'Donnell, T., Khurana, S., King, L.R., Manischewitz, J., Golding, H., Suphaphiphat, P., Carfi, A., Settembre, E.C., Dormitzer, P.R., Kepler, T.B., Zhang, R., Moody, M.A., Haynes, B.F., Liao, H.X., Shaw, D.E., Harrison, S.C.(2013) Proc Natl Acad Sci U S A 110: 264-269
- PubMed: 23175789
- DOI: https://doi.org/10.1073/pnas.1218256109
- Primary Citation of Related Structures:
4HK0, 4HK3, 4HKB, 4HKX - PubMed Abstract:
Affinity maturation refines a naive B-cell response by selecting mutations in antibody variable domains that enhance antigen binding. We describe a B-cell lineage expressing broadly neutralizing influenza virus antibodies derived from a subject immunized with the 2007 trivalent vaccine. The lineage comprises three mature antibodies, the unmutated common ancestor, and a common intermediate. Their heavy-chain complementarity determining region inserts into the conserved receptor-binding pocket of influenza HA. We show by analysis of structures, binding kinetics and long time-scale molecular dynamics simulations that antibody evolution in this lineage has rigidified the initially flexible heavy-chain complementarity determining region by two nearly independent pathways and that this preconfiguration accounts for most of the affinity gain. The results advance our understanding of strategies for developing more broadly effective influenza vaccines.
Organizational Affiliation:
Laboratory of Molecular Medicine, Children's Hospital, Harvard Medical School and Howard Hughes Medical Institute, Boston, MA 02115, USA.