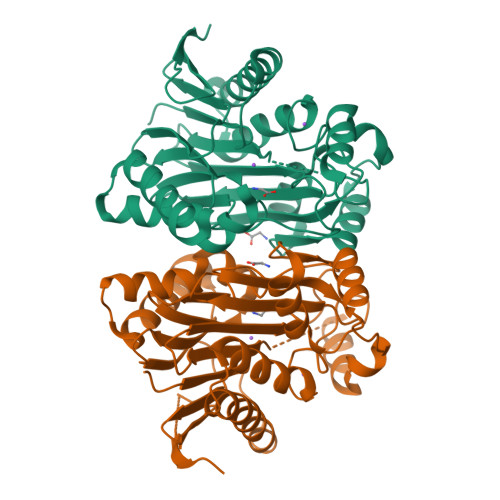
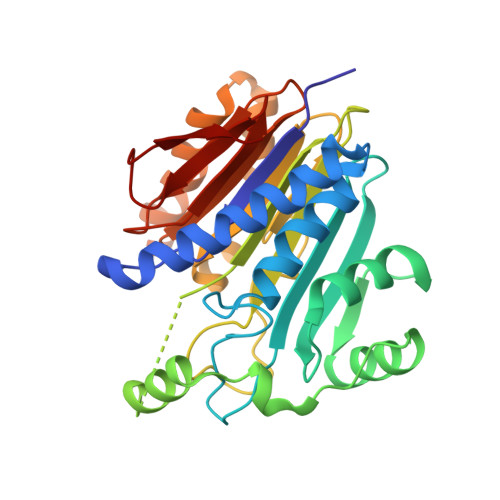
Elucidation of the Specific Function of the Conserved Threonine Triad Responsible for Human l-Asparaginase Autocleavage and Substrate Hydrolysis.
Nomme, J., Su, Y., Lavie, A.(2014) J Mol Biology 426: 2471-2485
- PubMed: 24768817
- DOI: https://doi.org/10.1016/j.jmb.2014.04.016
- Primary Citation of Related Structures:
4O0C, 4O0D, 4O0E, 4O0F, 4O0G, 4O0H - PubMed Abstract:
Our long-term goal is the design of a human l-asparaginase (hASNase3) variant, suitable for use in cancer therapy without the immunogenicity problems associated with the currently used bacterial enzymes. Asparaginases catalyze the hydrolysis of the amino acid asparagine to aspartate and ammonia. The key property allowing for the depletion of blood asparagine by bacterial asparaginases is their low micromolar KM value. In contrast, human enzymes have a millimolar KM for asparagine. Toward the goal of engineering an hASNase3 variant with micromolar KM, we conducted a structure/function analysis of the conserved catalytic threonine triad of this human enzyme. As a member of the N-terminal nucleophile family, to become enzymatically active, hASNase3 must undergo autocleavage between residues Gly167 and Thr168. To determine the individual contribution of each of the three conserved active-site threonines (threonine triad Thr168, Thr186, Thr219) for the enzyme-activating autocleavage and asparaginase reactions, we prepared the T168S, T186V and T219A/V mutants. These mutants were tested for their ability to cleave and to catalyze asparagine hydrolysis, in addition to being examined structurally. We also elucidated the first N-terminal nucleophile plant-type asparaginase structure in the covalent intermediate state. Our studies indicate that, while not all triad threonines are required for the cleavage reaction, all are essential for the asparaginase activity. The increased understanding of hASNase3 function resulting from these studies reveals the key regions that govern cleavage and the asparaginase reaction, which may inform the design of variants that attain a low KM for asparagine.
Organizational Affiliation:
Department of Biochemistry and Molecular Genetics, University of Illinois at Chicago, Chicago, IL 60607, USA.