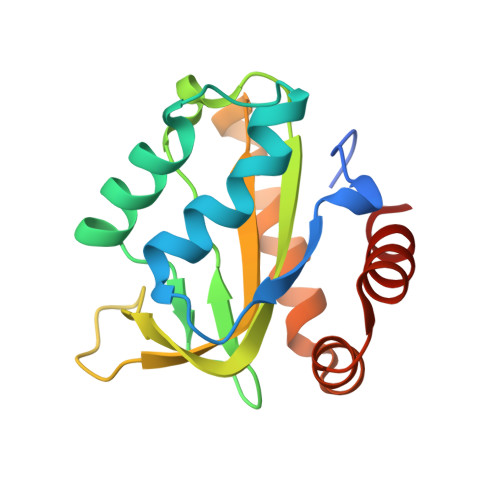
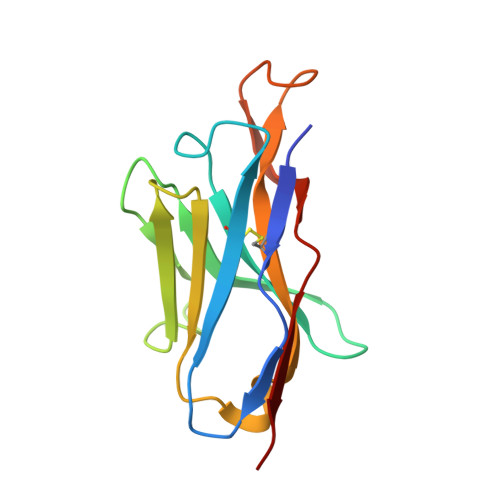
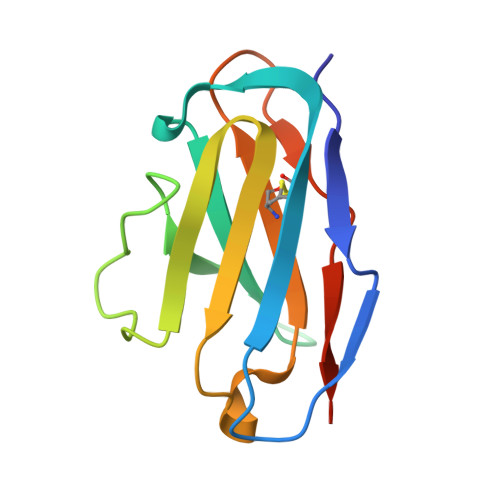
Ontogeny of Recognition Specificity and Functionality for the Broadly Neutralizing Anti-HIV Antibody 4E10.
Finton, K.A., Friend, D., Jaffe, J., Gewe, M., Holmes, M.A., Larman, H.B., Stuart, A., Larimore, K., Greenberg, P.D., Elledge, S.J., Stamatatos, L., Strong, R.K.(2014) PLoS Pathog 10: e1004403-e1004403
- PubMed: 25254371
- DOI: https://doi.org/10.1371/journal.ppat.1004403
- Primary Citation of Related Structures:
4LRN, 4M62, 4M8Q, 4OB5, 4ODX - PubMed Abstract:
The process of antibody ontogeny typically improves affinity, on-rate, and thermostability, narrows polyspecificity, and rigidifies the combining site to the conformer optimal for binding from the broader ensemble accessible to the precursor. However, many broadly-neutralizing anti-HIV antibodies incorporate unusual structural elements and recognition specificities or properties that often lead to autoreactivity. The ontogeny of 4E10, an autoreactive antibody with unexpected combining site flexibility, was delineated through structural and biophysical comparisons of the mature antibody with multiple potential precursors. 4E10 gained affinity primarily by off-rate enhancement through a small number of mutations to a highly conserved recognition surface. Controverting the conventional paradigm, the combining site gained flexibility and autoreactivity during ontogeny, while losing thermostability, though polyspecificity was unaffected. Details of the recognition mechanism, including inferred global effects due to 4E10 binding, suggest that neutralization by 4E10 may involve mechanisms beyond simply binding, also requiring the ability of the antibody to induce conformational changes distant from its binding site. 4E10 is, therefore, unlikely to be re-elicited by conventional vaccination strategies.
Organizational Affiliation:
Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, Washington, United States of America.