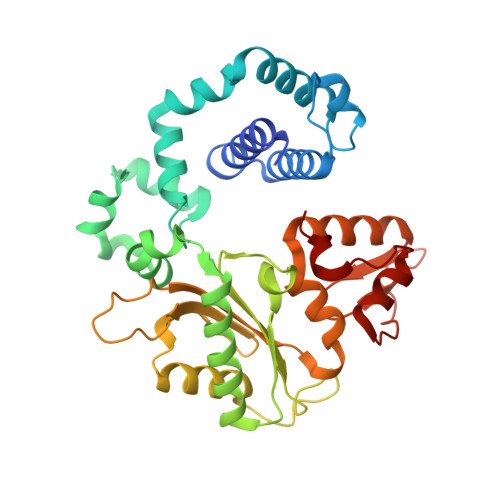
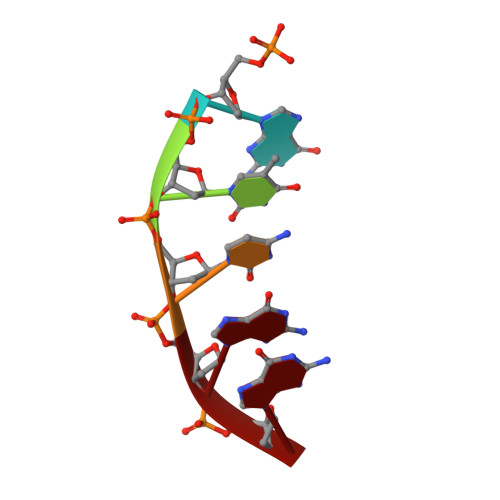
Uncovering the polymerase-induced cytotoxicity of an oxidized nucleotide.
Freudenthal, B.D., Beard, W.A., Perera, L., Shock, D.D., Kim, T., Schlick, T., Wilson, S.H.(2015) Nature 517: 635-639
- PubMed: 25409153
- DOI: https://doi.org/10.1038/nature13886
- PubMed Abstract:
Oxidative stress promotes genomic instability and human diseases. A common oxidized nucleoside is 8-oxo-7,8-dihydro-2'-deoxyguanosine, which is found both in DNA (8-oxo-G) and as a free nucleotide (8-oxo-dGTP). Nucleotide pools are especially vulnerable to oxidative damage. Therefore cells encode an enzyme (MutT/MTH1) that removes free oxidized nucleotides. This cleansing function is required for cancer cell survival and to modulate Escherichia coli antibiotic sensitivity in a DNA polymerase (pol)-dependent manner. How polymerases discriminate between damaged and non-damaged nucleotides is not well understood. This analysis is essential given the role of oxidized nucleotides in mutagenesis, cancer therapeutics, and bacterial antibiotics. Even with cellular sanitizing activities, nucleotide pools contain enough 8-oxo-dGTP to promote mutagenesis. This arises from the dual coding potential where 8-oxo-dGTP(anti) base pairs with cytosine and 8-oxo-dGTP(syn) uses its Hoogsteen edge to base pair with adenine. Here we use time-lapse crystallography to follow 8-oxo-dGTP insertion opposite adenine or cytosine with human pol β, to reveal that insertion is accommodated in either the syn- or anti-conformation, respectively. For 8-oxo-dGTP(anti) insertion, a novel divalent metal relieves repulsive interactions between the adducted guanine base and the triphosphate of the oxidized nucleotide. With either templating base, hydrogen-bonding interactions between the bases are lost as the enzyme reopens after catalysis, leading to a cytotoxic nicked DNA repair intermediate. Combining structural snapshots with kinetic and computational analysis reveals how 8-oxo-dGTP uses charge modulation during insertion that can lead to a blocked DNA repair intermediate.
Organizational Affiliation:
Laboratory of Structural Biology, National Institute of Environmental Health Sciences, National Institutes of Health, PO Box 12233, Research Triangle Park, North Carolina 27709-2233, USA.