Discovery of (2S)-8-[(3R)-3-Methylmorpholin-4-Yl]-1-(3-Methyl-2-Oxobutyl)-2-(Trifluoromethyl)-3,4-Dihydro-2H-Pyrimido[1,2-A]Pyrimidin-6-One: A Novel Potent and Selective Inhibitor of Vps34 for the Treatment of Solid Tumors.
Pasquier, B., El-Ahmad, Y., Filoche-Romme, B., Dureuil, C., Fassy, F., Abecassis, P., Mathieu, M., Bertrand, T., Benard, T., Barriere, C., El Batti, S., Letallec, J., Sonnefraud, V., Brollo, M., Delbarre, L., Loyau, V., Pilorge, F., Bertin, L., Richepin, P., Arigon, J., Labrosse, J., Clement, J., Durand, F., Combet, R., Perraut, P., Leroy, V., Gay, F., Lefrancois, D., Bretin, F., Marquette, J., Michot, N., Caron, A., Castell, C., Schio, L., Mccort, G., Goulaouic, H., Garcia-Echeverria, C., Ronan, B.(2015) J Med Chem 58: 376
- PubMed: 25402320
- DOI: https://doi.org/10.1021/jm5013352
- Primary Citation of Related Structures:
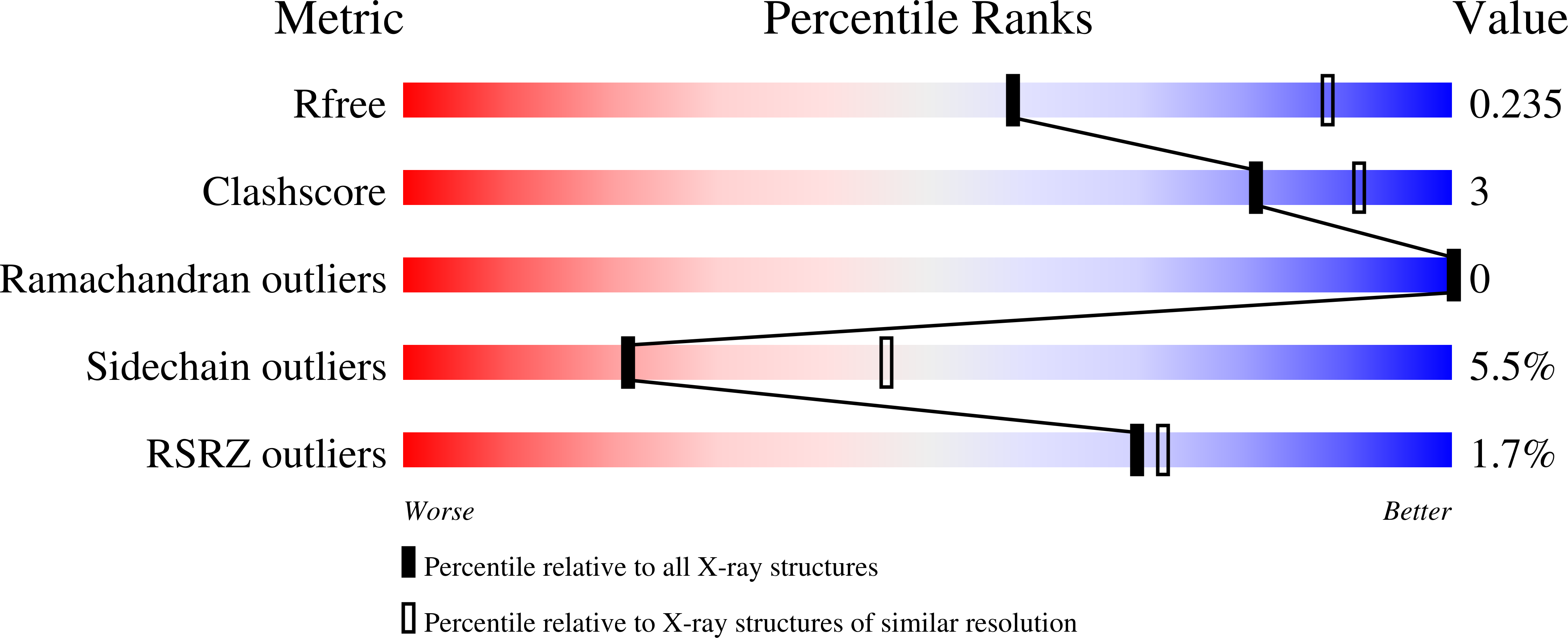
4UWF, 4UWG, 4UWH, 4UWK, 4UWL - PubMed Abstract:
Vps34 (the human class III phosphoinositide 3-kinase) is a lipid kinase involved in vesicle trafficking and autophagy and therefore constitutes an interesting target for cancer treatment. Because of the lack of specific Vps34 kinase inhibitors, we aimed to identify such compounds to further validate the role of this lipid kinase in cancer maintenance and progression. Herein, we report the discovery of a series of tetrahydropyrimidopyrimidinone derivatives. Starting with hit compound 1a, medicinal chemistry optimization led to compound 31. This molecule displays potent activity, an exquisite selectivity for Vps34 with excellent properties. The X-ray crystal structure of compound 31 in human Vps34 illustrates how the unique molecular features of the morpholine synthon bestows selectivity against class I PI3Ks. This molecule exhibits suitable in vivo mouse PK parameters and induces a sustained inhibition of Vps34 upon acute administration. Compound 31 constitutes an optimized Vps34 inhibitor that could be used to investigate human cancer biology.
Organizational Affiliation:
Oncology Drug Discovery, ‡Drug Disposition and Safety, §Structure Design Informatics and Structural Biology, ∥Pharmaceutical Sciences Operations, ⊥Protein Production, and #Global Biotherapeutic, Sanofi , 13 Quai Jules Guesde, 94403 Vitry-sur-Seine, France.