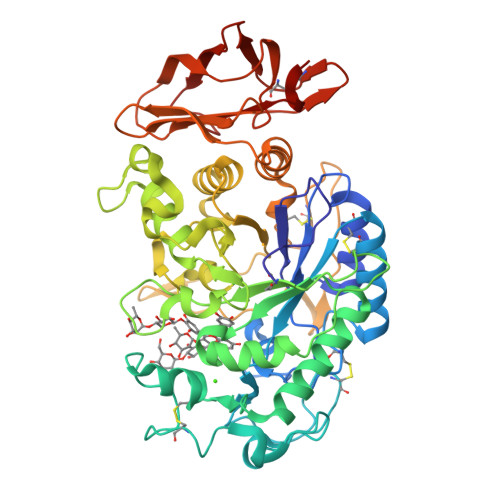
The amylase inhibitor montbretin A reveals a new glycosidase inhibition motif.
Williams, L.K., Zhang, X., Caner, S., Tysoe, C., Nguyen, N.T., Wicki, J., Williams, D.E., Coleman, J., McNeill, J.H., Yuen, V., Andersen, R.J., Withers, S.G., Brayer, G.D.(2015) Nat Chem Biol 11: 691-696
- PubMed: 26214255
- DOI: https://doi.org/10.1038/nchembio.1865
- Primary Citation of Related Structures:
4W93 - PubMed Abstract:
The complex plant flavonol glycoside montbretin A is a potent (Ki = 8 nM) and specific inhibitor of human pancreatic α-amylase with potential as a therapeutic for diabetes and obesity. Controlled degradation studies on montbretin A, coupled with inhibition analyses, identified an essential high-affinity core structure comprising the myricetin and caffeic acid moieties linked via a disaccharide. X-ray structural analyses of the montbretin A-human α-amylase complex confirmed the importance of this core structure and revealed a novel mode of glycosidase inhibition wherein internal π-stacking interactions between the myricetin and caffeic acid organize their ring hydroxyls for optimal hydrogen bonding to the α-amylase catalytic residues D197 and E233. This novel inhibitory motif can be reproduced in a greatly simplified analog, offering potential for new strategies for glycosidase inhibition and therapeutic development.
Organizational Affiliation:
Department of Biochemistry and Molecular Biology, Faculty of Medicine, University of British Columbia, Vancouver, British Columbia, Canada.