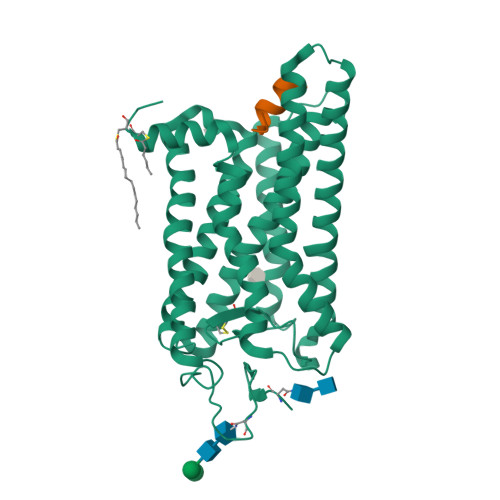
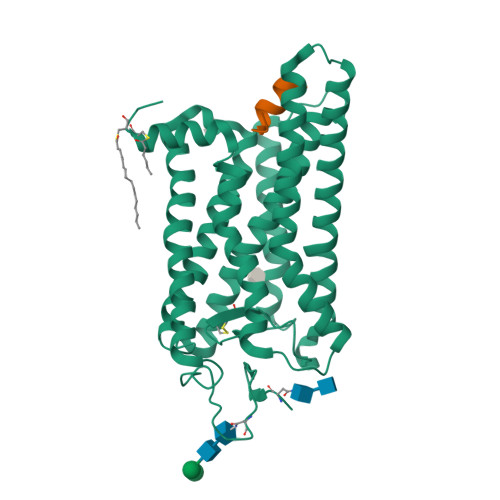
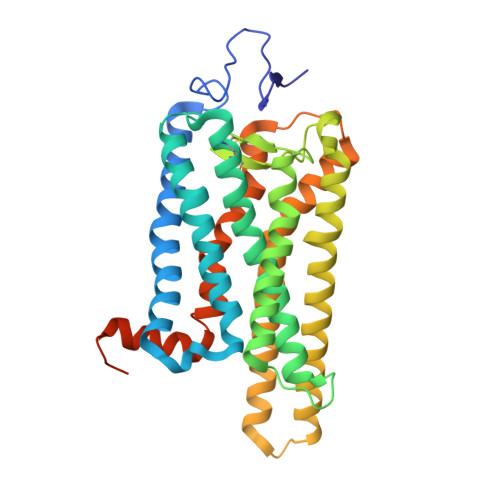
The High-Resolution Structure of Activated Opsin Reveals a Conserved Solvent Network in the Transmembrane Region Essential for Activation.
Blankenship, E., Vahedi-Faridi, A., Lodowski, D.T.(2015) Structure 23: 2358-2364
- PubMed: 26526852
- DOI: https://doi.org/10.1016/j.str.2015.09.015
- Primary Citation of Related Structures:
4X1H - PubMed Abstract:
Rhodopsin, a light-activated G protein coupled receptor (GPCR), has been the subject of numerous biochemical and structural investigations, serving as a model receptor for GPCRs and their activation. We present the 2.3-Å resolution structure of native source rhodopsin stabilized in a conformation competent for G protein binding. An extensive water-mediated hydrogen bond network linking the chromophore binding site to the site of G protein binding is observed, providing connections to conserved motifs essential for GPCR activation. Comparison of this extensive solvent-mediated hydrogen-bonding network with the positions of ordered solvent in earlier crystallographic structures of rhodopsin photointermediates reveals both static structural and dynamic functional water-protein interactions present during the activation process. When considered along with observations that solvent occupies similar positions in the structures of other GPCRs, these analyses strongly support an integral role for this dynamic ordered water network in both rhodopsin and GPCR activation.
Organizational Affiliation:
Department of Nutrition, Case Western Reserve University School of Medicine, 10900 Euclid Avenue, Cleveland, OH 44106, USA; Center for Proteomics and Bioinformatics, Case Western Reserve University School of Medicine, 10900 Euclid Avenue, Cleveland, OH 44106, USA.