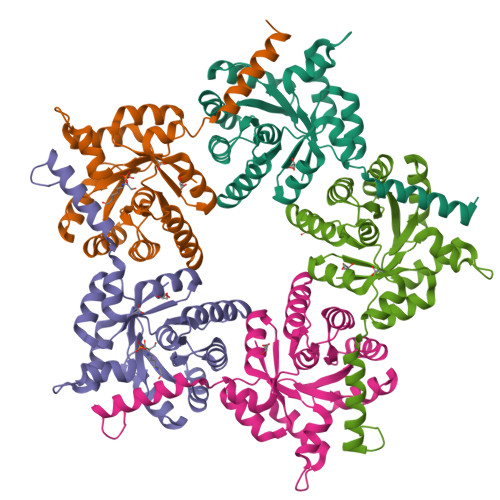
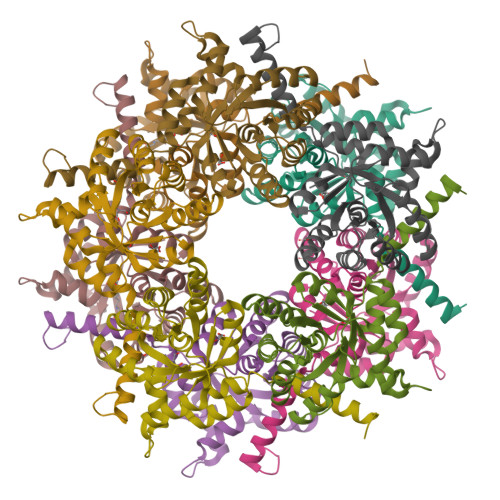
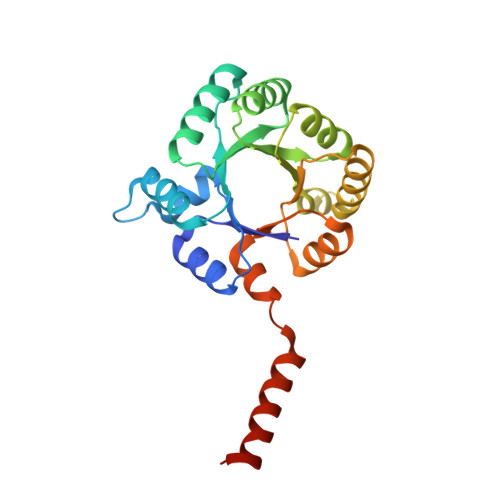
Converting Transaldolase into Aldolase through Swapping of the Multifunctional Acid-Base Catalyst: Common and Divergent Catalytic Principles in F6P Aldolase and Transaldolase.
Sautner, V., Friedrich, M.M., Lehwess-Litzmann, A., Tittmann, K.(2015) Biochemistry 54: 4475-4486
- PubMed: 26131847
- DOI: https://doi.org/10.1021/acs.biochem.5b00283
- Primary Citation of Related Structures:
4XZ9 - PubMed Abstract:
Transaldolase (TAL) and fructose-6-phosphate aldolase (FSA) both belong to the class I aldolase family and share a high degree of structural similarity and sequence identity. The molecular basis of the different reaction specificities (transferase vs aldolase) has remained enigmatic. A notable difference between the active sites is the presence of either a TAL-specific Glu (Gln in FSA) or a FSA-specific Tyr (Phe in TAL). Both residues seem to have analoguous multifunctional catalytic roles but are positioned at different faces of the substrate locale. We have engineered a TAL double variant (Glu to Gln and Phe to Tyr) with an active site resembling that of FSA. This variant indeed exhibits aldolase activity as its main activity with a catalytic efficiency even larger than that of authentic FSA, while TAL activity is greatly impaired. Structural analysis of this variant in complex with the dihydroxyacetone Schiff base formed upon substrate cleavage identifies the introduced Tyr (genuine in FSA) to catalyze protonation of the central carbanion-enamine intermediate as a key determinant of the aldolase reaction. Our studies pinpoint that the Glu in TAL and the Tyr in FSA, although located at different positions at the active site, similarly act as bona fide acid-base catalysts in numerous catalytic steps, including substrate binding, dehydration of the carbinolamine, and substrate cleavage. We propose that the different spatial positions of the multifunctional Glu in TAL and of the corresponding multifunctional Tyr in FSA relative to the substrate locale are critically controlling reaction specificity through either unfavorable (TAL) or favorable (FSA) geometry of proton transfer onto the common carbanion-enamine intermediate. The presence of both potential acid-base residues, Glu and Tyr, in the active site of TAL has deleterious effects on substrate binding and cleavage, most likely resulting from a differently organized H-bonding network. Large-scale motions of the protein associated with opening and closing of the active site that seem to bear relevance for catalysis are observed as covalent intermediates are exclusively observed in the "closed" conformation of the active site. Pre-steady-state kinetics are used to monitor catalytic processes and structural transitions and to refine the kinetic framework of TAL catalysis.
Organizational Affiliation:
Göttingen Center for Molecular Biosciences, Department of Molecular Enzymology, Georg-August University Göttingen, Ernst-Caspari-Haus, Justus-von-Liebig-Weg 11, D-37077 Göttingen, Germany.