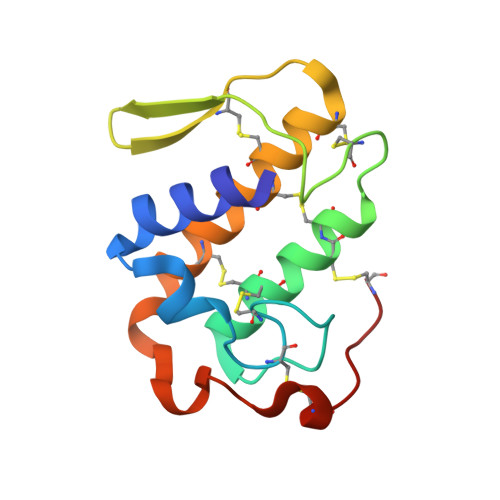
Structural Basis for the Inhibition of a Phospholipase A2-Like Toxin by Caffeic and Aristolochic Acids.
Fernandes, C.A., Cardoso, F.F., Cavalcante, W.G., Soares, A.M., Dal-Pai, M., Gallacci, M., Fontes, M.R.(2015) PLoS One 10: e0133370-e0133370
- PubMed: 26192963
- DOI: https://doi.org/10.1371/journal.pone.0133370
- Primary Citation of Related Structures:
4YU7, 4YZ7 - PubMed Abstract:
One of the main challenges in toxicology today is to develop therapeutic alternatives for the treatment of snake venom injuries that are not efficiently neutralized by conventional serum therapy. Venom phospholipases A2 (PLA2s) and PLA2-like proteins play a fundamental role in skeletal muscle necrosis, which can result in permanent sequelae and disability. This leads to economic and social problems, especially in developing countries. In this work, we performed structural and functional studies with Piratoxin-I, a Lys49-PLA2 from Bothropspirajai venom, complexed with two compounds present in several plants used in folk medicine against snakebites. These ligands partially neutralized the myotoxic activity of PrTX-I towards binding on the two independent sites of interaction between Lys49-PLA2 and muscle membrane. Our results corroborate the previously proposed mechanism of action of PLA2s-like and provide insights for the design of structure-based inhibitors that could prevent the permanent injuries caused by these proteins in snakebite victims.
Organizational Affiliation:
Dep. de Física e Biofísica, Instituto de Biociências, UNESP-Universidade Estadual Paulista, Botucatu, São Paulo, Brazil; Instituto Nacional de Ciência e Tecnologia em Toxinas, CNPq, São Paulo, São Paulo, Brazil.