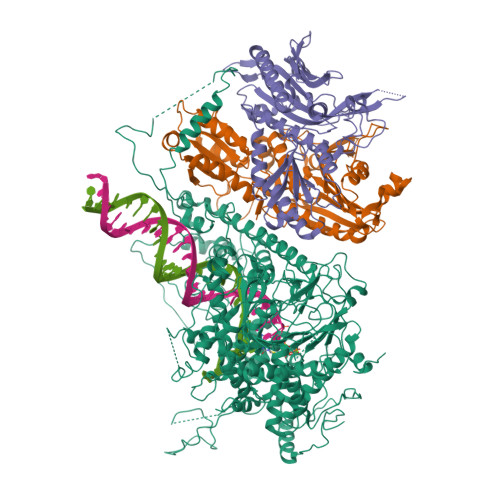
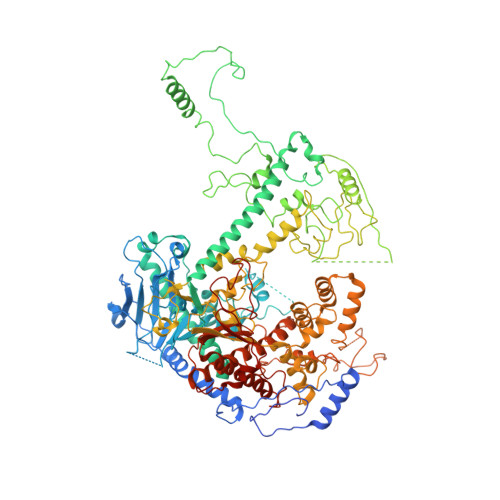
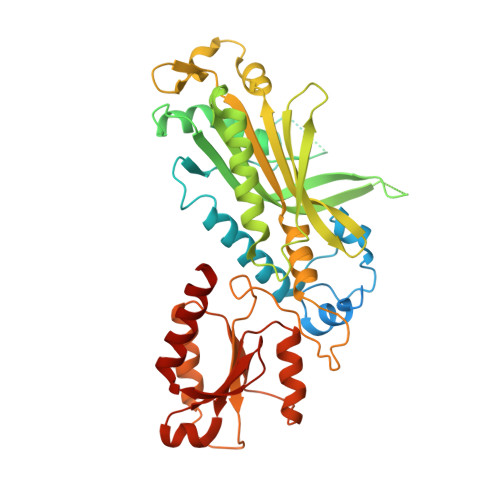
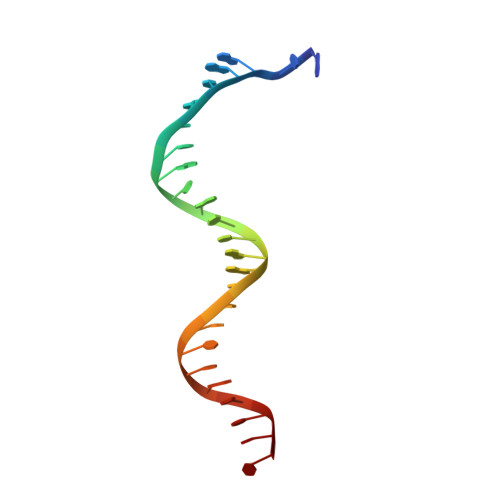
Probing the structural and molecular basis of nucleotide selectivity by human mitochondrial DNA polymerase gamma.
Sohl, C.D., Szymanski, M.R., Mislak, A.C., Shumate, C.K., Amiralaei, S., Schinazi, R.F., Anderson, K.S., Yin, Y.W.(2015) Proc Natl Acad Sci U S A 112: 8596-8601
- PubMed: 26124101
- DOI: https://doi.org/10.1073/pnas.1421733112
- Primary Citation of Related Structures:
5C51, 5C52, 5C53 - PubMed Abstract:
Nucleoside analog reverse transcriptase inhibitors (NRTIs) are the essential components of highly active antiretroviral (HAART) therapy targeting HIV reverse transcriptase (RT). NRTI triphosphates (NRTI-TP), the biologically active forms, act as chain terminators of viral DNA synthesis. Unfortunately, NRTIs also inhibit human mitochondrial DNA polymerase (Pol γ), causing unwanted mitochondrial toxicity. Understanding the structural and mechanistic differences between Pol γ and RT in response to NRTIs will provide invaluable insight to aid in designing more effective drugs with lower toxicity. The NRTIs emtricitabine [(-)-2,3'-dideoxy-5-fluoro-3'-thiacytidine, (-)-FTC] and lamivudine, [(-)-2,3'-dideoxy-3'-thiacytidine, (-)-3TC] are both potent RT inhibitors, but Pol γ discriminates against (-)-FTC-TP by two orders of magnitude better than (-)-3TC-TP. Furthermore, although (-)-FTC-TP is only slightly more potent against HIV RT than its enantiomer (+)-FTC-TP, it is discriminated by human Pol γ four orders of magnitude more efficiently than (+)-FTC-TP. As a result, (-)-FTC is a much less toxic NRTI. Here, we present the structural and kinetic basis for this striking difference by identifying the discriminator residues of drug selectivity in both viral and human enzymes responsible for substrate selection and inhibitor specificity. For the first time, to our knowledge, this work illuminates the mechanism of (-)-FTC-TP differential selectivity and provides a structural scaffold for development of novel NRTIs with lower toxicity.
Organizational Affiliation:
Department of Pharmacology, Yale University School of Medicine, New Haven, CT 06520;