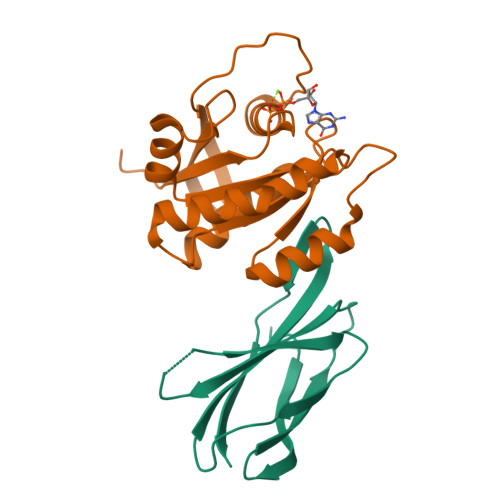
Inhibition of RAS function through targeting an allosteric regulatory site.
Spencer-Smith, R., Koide, A., Zhou, Y., Eguchi, R.R., Sha, F., Gajwani, P., Santana, D., Gupta, A., Jacobs, M., Herrero-Garcia, E., Cobbert, J., Lavoie, H., Smith, M., Rajakulendran, T., Dowdell, E., Okur, M.N., Dementieva, I., Sicheri, F., Therrien, M., Hancock, J.F., Ikura, M., Koide, S., O'Bryan, J.P.(2017) Nat Chem Biol 13: 62-68
- PubMed: 27820802
- DOI: https://doi.org/10.1038/nchembio.2231
- Primary Citation of Related Structures:
5E95 - PubMed Abstract:
RAS GTPases are important mediators of oncogenesis in humans. However, pharmacological inhibition of RAS has proved challenging. Here we describe a functionally critical region, located outside the effector lobe of RAS, that can be targeted for inhibition. We developed NS1, a synthetic binding protein (monobody) that bound with high affinity to both GTP- and GDP-bound states of H-RAS and K-RAS but not N-RAS. NS1 potently inhibited growth factor signaling and oncogenic H-RAS- and K-RAS-mediated signaling and transformation but did not block oncogenic N-RAS, BRAF or MEK1. NS1 bound the α4-β6-α5 region of RAS, which disrupted RAS dimerization and nanoclustering and led to blocking of CRAF-BRAF heterodimerization and activation. These results establish the importance of the α4-β6-α5 interface in RAS-mediated signaling and define a previously unrecognized site in RAS for inhibiting RAS function.
Organizational Affiliation:
Department of Pharmacology, University of Illinois at Chicago, Chicago, Illinois, USA.