Structure-function analysis for the hydroxylation of Delta 4 C21-steroids by the myxobacterial CYP260B1.
Salamanca-Pinzon, S.G., Khatri, Y., Carius, Y., Keller, L., Muller, R., Lancaster, C.R., Bernhardt, R.(2016) FEBS Lett 590: 1838-1851
- PubMed: 27177597
- DOI: https://doi.org/10.1002/1873-3468.12217
- Primary Citation of Related Structures:
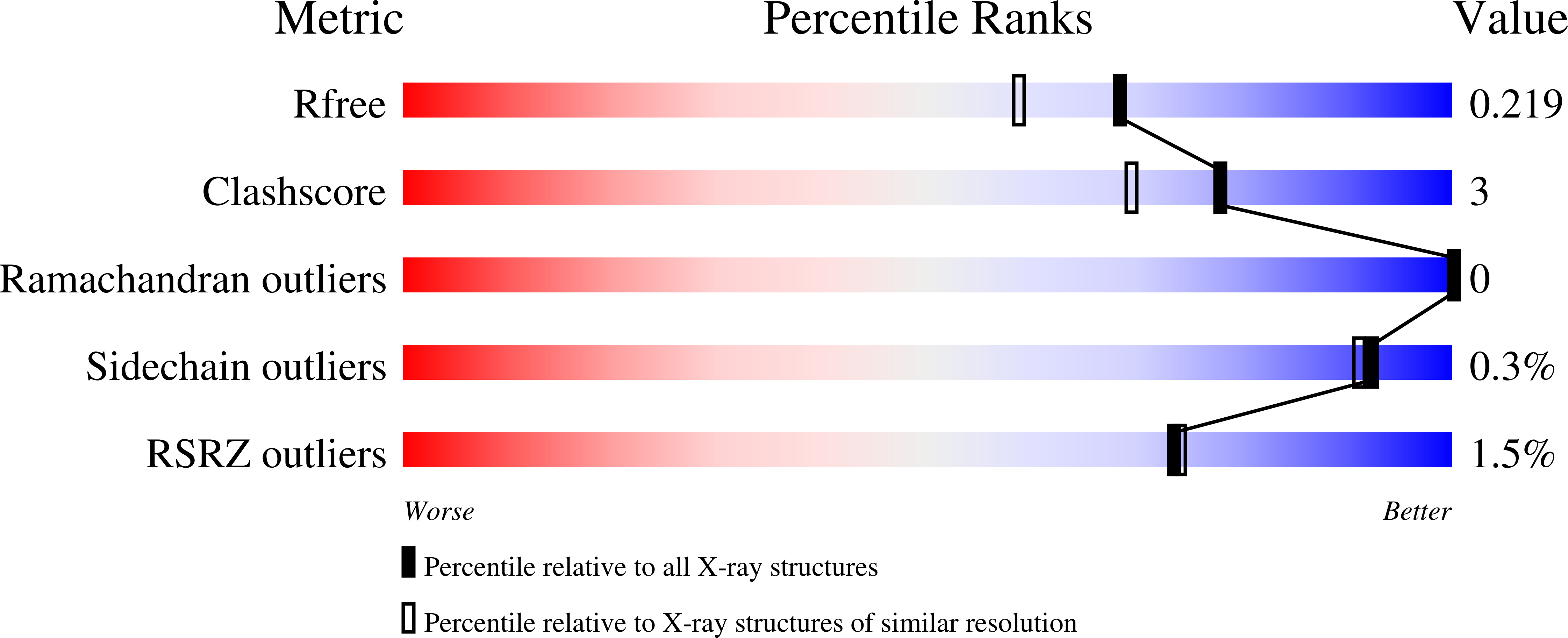
5HIW - PubMed Abstract:
Myxobacterial CYP260B1 from Sorangium cellulosum was heterologously expressed in Escherichia coli and purified. The in vitro conversion of a small focused substrate library comprised of Δ4 C21-steroids and steroidal drugs using surrogate bovine redox partners shows that CYP260B1 is a novel steroid hydroxylase. CYP260B1 performs the regio- and stereoselective hydroxylation of the glucocorticoid cortodoxone (RSS) to produce 6β-OH-RSS. The substrate-free crystal structure of CYP260B1 (PDB 5HIW) was resolved. Docking of the tested ligands into the crystal structure suggested that the C17 hydroxy moiety and the presence of either a keto or a hydroxy group at C11 determine the selectivity of hydroxylation.
Organizational Affiliation:
Institute of Biochemistry, Saarland University, Saarbrücken, Germany.