PPARgamma-RXRalpha(S427F) heterodimer in complex with SRC-1, rosiglitazone, and 9-cis-retanoic acid
Korpal, M., Zhu, P., Bloudoff, K., Larsen, N.A., Fekkes, P.To be published.
Experimental Data Snapshot
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Retinoic acid receptor RXR-alpha | 242 | Homo sapiens | Mutation(s): 1 Gene Names: RXRA, NR2B1 | 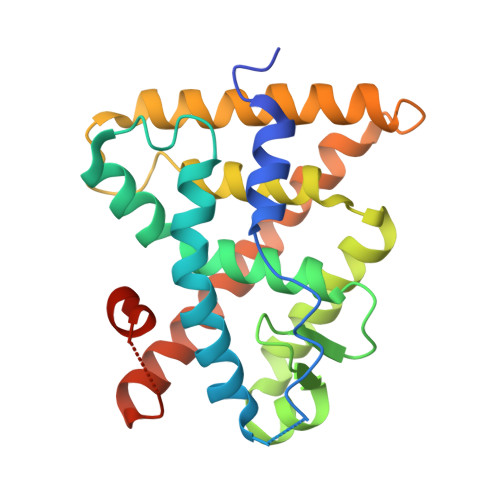 | |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P19793 (Homo sapiens) Explore P19793 Go to UniProtKB: P19793 | |||||
PHAROS: P19793 GTEx: ENSG00000186350 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P19793 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Peroxisome proliferator-activated receptor gamma | B [auth D] | 273 | Homo sapiens | Mutation(s): 0 Gene Names: PPARG, NR1C3 | 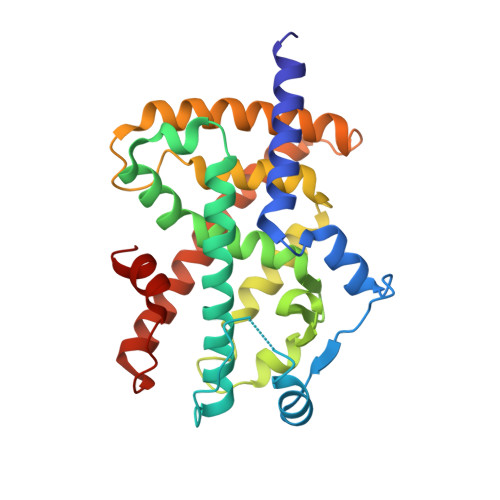 |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for P37231 (Homo sapiens) Explore P37231 Go to UniProtKB: P37231 | |||||
PHAROS: P37231 GTEx: ENSG00000132170 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P37231 | ||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 3 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| Nuclear receptor coactivator 1 | C [auth E], D [auth F] | 27 | Homo sapiens | Mutation(s): 2 EC: 2.3.1.48 |  |
UniProt & NIH Common Fund Data Resources | |||||
Find proteins for Q15788 (Homo sapiens) Explore Q15788 Go to UniProtKB: Q15788 | |||||
PHAROS: Q15788 GTEx: ENSG00000084676 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | Q15788 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| BRL Query on BRL | F [auth D] | 2,4-THIAZOLIDIINEDIONE, 5-[[4-[2-(METHYL-2-PYRIDINYLAMINO)ETHOXY]PHENYL]METHYL]-(9CL) C18 H19 N3 O3 S YASAKCUCGLMORW-HNNXBMFYSA-N |  | ||
| 9CR Query on 9CR | E [auth A] | (9cis)-retinoic acid C20 H28 O2 SHGAZHPCJJPHSC-ZVCIMWCZSA-N |  | ||
| Binding Affinity Annotations | |||
|---|---|---|---|
| ID | Source | Binding Affinity | |
| 9CR | BindingDB: 5JI0 | Ki: min: 3.8, max: 583 (nM) from 12 assay(s) | |
| Kd: min: 1.5, max: 1810 (nM) from 20 assay(s) | |||
| IC50: min: 4, max: 82 (nM) from 5 assay(s) | |||
| EC50: min: 1.5, max: 316 (nM) from 25 assay(s) | |||
| BRL | BindingDB: 5JI0 | Ki: min: 8, max: 59 (nM) from 7 assay(s) | |
| Kd: min: 1.2, max: 3300 (nM) from 3 assay(s) | |||
| IC50: min: 7.7, max: 6.00e+4 (nM) from 14 assay(s) | |||
| EC50: min: 1, max: 1600 (nM) from 47 assay(s) | |||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 53.88 | α = 90 |
| b = 66.527 | β = 90 |
| c = 165.927 | γ = 90 |
| Software Name | Purpose |
|---|---|
| REFMAC | refinement |
| HKL-2000 | data reduction |
| HKL-2000 | data scaling |
| MOLREP | phasing |