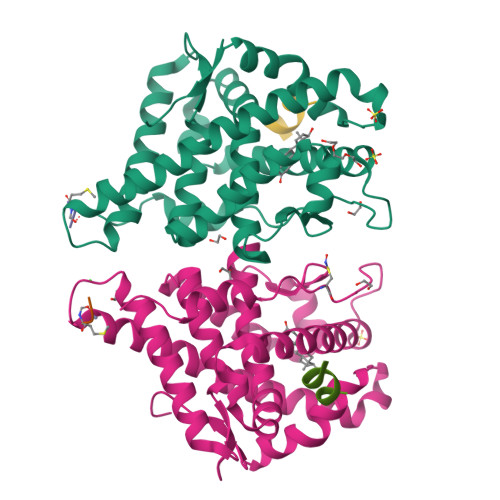
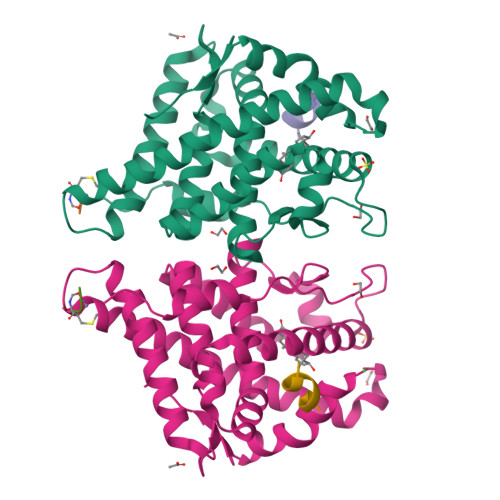
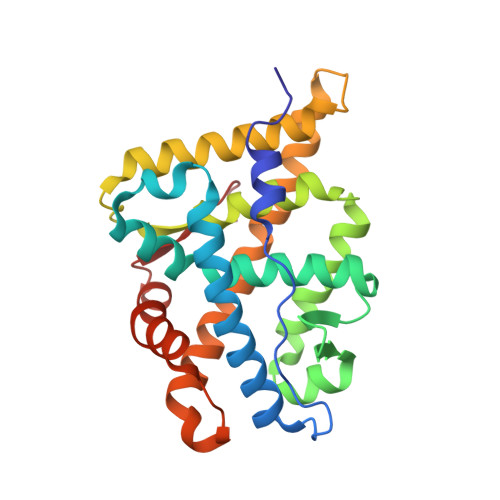
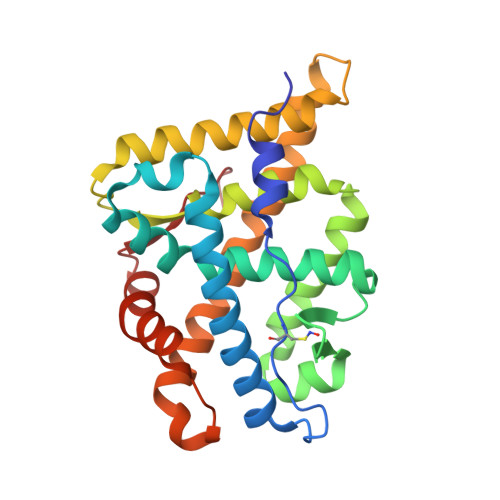
Structure of the homodimeric androgen receptor ligand-binding domain.
Nadal, M., Prekovic, S., Gallastegui, N., Helsen, C., Abella, M., Zielinska, K., Gay, M., Vilaseca, M., Taules, M., Houtsmuller, A.B., van Royen, M.E., Claessens, F., Fuentes-Prior, P., Estebanez-Perpina, E.(2017) Nat Commun 8: 14388-14388
- PubMed: 28165461
- DOI: https://doi.org/10.1038/ncomms14388
- Primary Citation of Related Structures:
5JJM - PubMed Abstract:
The androgen receptor (AR) plays a crucial role in normal physiology, development and metabolism as well as in the aetiology and treatment of diverse pathologies such as androgen insensitivity syndromes (AIS), male infertility and prostate cancer (PCa). Here we show that dimerization of AR ligand-binding domain (LBD) is induced by receptor agonists but not by antagonists. The 2.15-Å crystal structure of homodimeric, agonist- and coactivator peptide-bound AR-LBD unveils a 1,000-Å 2 large dimerization surface, which harbours over 40 previously unexplained AIS- and PCa-associated point mutations. An AIS mutation in the self-association interface (P767A) disrupts dimer formation in vivo, and has a detrimental effect on the transactivating properties of full-length AR, despite retained hormone-binding capacity. The conservation of essential residues suggests that the unveiled dimerization mechanism might be shared by other nuclear receptors. Our work defines AR-LBD homodimerization as an essential step in the proper functioning of this important transcription factor.
Organizational Affiliation:
Department of Biochemistry and Molecular Biomedicine, Institute of Biomedicine (IBUB) of the University of Barcelona (UB), Barcelona 08028, Spain.