Protein Recognition by Functionalized Sulfonatocalix[4]arenes.
Doolan, A.M., Rennie, M.L., Crowley, P.B.(2018) Chemistry 24: 984-991
- PubMed: 29125201
- DOI: https://doi.org/10.1002/chem.201704931
- Primary Citation of Related Structures:
5KPF, 5LFT, 5T8W - PubMed Abstract:
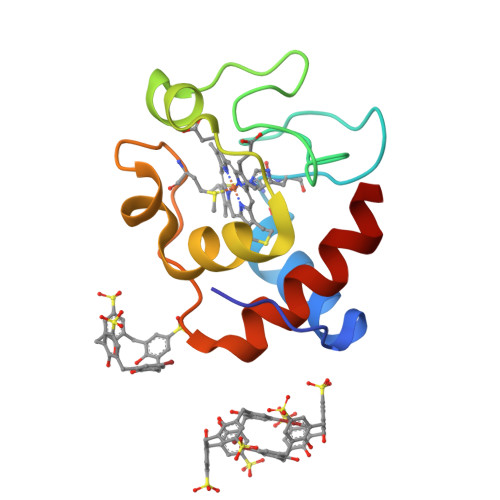
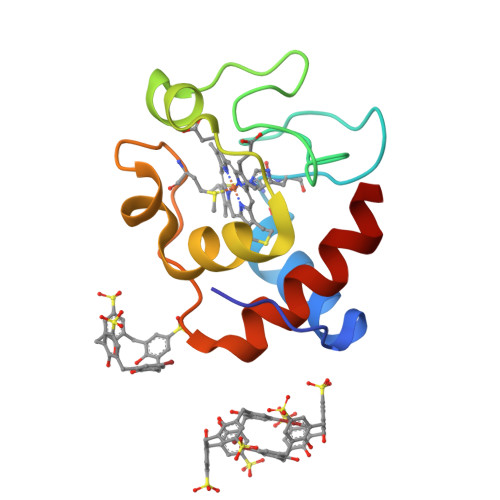
The interactions of two mono-functionalized sulfonatocalix[4]arenes with cytochrome c were investigated by structural and thermodynamic methods. The replacement of a single sulfonate with either a bromo or a phenyl substituent resulted in altered recognition of cytochrome c as evidenced by X-ray crystallography. The bromo-substituted ligand yielded a new binding mode in which a self-encapsulated calixarene dimer contributed to crystal packing. This ligand also formed a weak halogen bond with the protein. The phenyl-substituted ligand was bound to Lys4 of cytochrome c, in a 1.7 Å resolution crystal structure. A dimeric packing arrangement mediated by ligand-ligand contacts in the crystal suggested a possible assembly mechanism. The different protein recognition properties of these calixarenes are discussed.
Organizational Affiliation:
School of Chemistry, National University of Ireland Galway, University Road, Galway, Ireland.